* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download MAT
X-inactivation wikipedia , lookup
Ridge (biology) wikipedia , lookup
Cell culture wikipedia , lookup
Genomic imprinting wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Secreted frizzled-related protein 1 wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Gene expression profiling wikipedia , lookup
Paracrine signalling wikipedia , lookup
Biochemical cascade wikipedia , lookup
List of types of proteins wikipedia , lookup
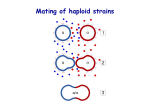
• Lecture 2: Read about the yeast MAT locus in Molecular Biology of the Gene. Watson et al. Chapter 10. Plus section on yeast as a model system • Read chapter 22 and chapter 10 [section on MATing type gene expression] in Molecular Cell Biology by Lodish et al [http://www.ncbi.nlm.nih.gov/books/bv.fcgi?rid=mcb] here it is section 14.1] Yeasts of opposite mating types can MATE • • • • • • Yeast cells can proliferate both as haploids (1n) and as diploids (2n); 2n cells are 1.2-fold bigger Haploid cells have one of two mating types: a or alpha Two haploid cells can mate to form a zygote; since yeast cannot move, cells must grow towards each other (shmoos) The diploid zygote starts dividing from the junction Under nitrogen starvation diploid cells undergo meiosis and sporulation to form an ascus with four haploid spores Thus, although yeast is unicellular, we can distinguish different cell types with different genetic programmes: – Haploid MATa versus MATalpha – Haploid versus Diploid (MATa/alpha) – Spores – Mothers and daughters Mating • Haploid cells produce specific mating factors, which bind to specific cellsurface receptors • Changes gene expression • Induces cell fusion, which produces a diploid cell Yeast sexual cycle • Caught in the act: cell attachment, cell fusion and nuclear fusion in an electron micrograph • Haploid cells produce mating peptide pheromones, i.e. a-factor and alpha-factor, to which the mating partner responds to prepare for mating Budding and Schmooed cells (A) Cells of Saccharomyces cerevisiae are usually spherical. (B) They become polarized when treated with mating factor from cells of the opposite mating type. The polarized cells are called “shmoos.” (C) Al Capp's famous cartoon character, the original Shmoo. (A and B, courtesy of Michael Snyder; C, © 1948 Capp Enterprises, Inc., all rights reserved.) Yeast sexual cycle • Central to sexual communication is the pheromone response pathway • This pathway is a complex system that controls the response of yeast cells to a- or alpha-factor • All blocks of that pathway consist of components conserved from yeast to human • The pathway consists of a specific pheromone receptor, that binds aor alpha-factor • Binding stimulates a signalling pathway that causes cell cycle arrest and coordinates expression of genes important for mating • The pathway is also needed to orient the cell towards its mating Pheromone-induced signalling Cytoskeleton changes to all cell fusion Transcriptional changes to promote mating Mating-type (MAT) locus A major genetic determinant of mating is the MAT LOCUS situated on chromosome III Two alleles that determine mating type • MAT • MATa How do these alleles determine mating type? Mating-type allele switching • Mating type is determined by which allele (a or ) is in the MAT locus • Mating-type specific allele (a or ) is introduced into MAT by recombination • Silent (not transcribed) copies of each allele are “stored” at HML and HMRa Genetic determination of yeast cell type • • • • The mating type locus encodes regulatory proteins, i.e. transcription factors The MATa locus encodes the a1 transcriptional activator (a2 has no known function) The a2 null also shows no growth, mating or sporulation defect The MATalpha locus encodes the alpha1 activator and the alpha2 repressor The mating type locus functions as a master regulator locus: it controls expression of many genes Transcription factors • MAT locus alleles encodes cell-specific transcription factors – MAT encodes Mat 1 and Mat 2 – MATa encodes Mata1 • MCM1 encodes a general transcription factor found in haploid and diploid cells. Regulation of cell-specific genes • Mat 1 (and Mcm1) activate -specific genes • Mat 2 (and Mcm1) repress a-specific genes • Mata1 and Mat 2 repress haploidspecific genes (no role for Mata1in haploid) cell Mat 1 and Mat 2 produced • Mat 1 and Mcm1 activate -specific genes • Mat 2 and Mcm1 repress a-specific genes a cell Mata1 produced • Mcm1 activates a-specific and haploidspecific genes a/ cell (diploid) • Mat 2 and Mcm1 repress a-specific genes • Mata1 and Mat 2 repress haploidspecific genes and repress MAT 1 Cell-specific genes • -specific genes are those needed to produce alpha-factor and the gene for the afactor receptor • a-specific genes are those needed for afactor production and the gene for the alphafactor receptor. • Haploid-specific genes include the RME gene encoding the meiosis repressor and the HO endonuclease (which catalyzes switching) • and those that encode proteins involved in the response to pheromone Some Transcription factors can act as activators or repressors • • • MCM1 is a yeast transcriptional activator with a DNA-binding domain shared by other members of the MADS-box family, including SRF. MCM1 binds cooperatively with the MATalpha1 protein to activate transcription of the alpha-specific genes, or with the MATalpha2 protein to repress transcription of the aspecific genes. Haploids can switch mating type! • • • • • Haploid cells can switch their mating type, i.e. from a to alpha or from alpha to a This is due to two silent mating type loci on the same chromosome, which become activated when translocated to the MAT locus The translocation is a gene conversion event initiated by the HO nuclease The switch ensures that in Nature yeast cells return quickly to the diploid, more stable stage Laboratory yeast strains lack the HO nuclease and hence have stable haploid phases Mating Type Switching in Yeast HO endonuclease • Mating-type switching involves recombination of a transcriptionally silent allele into a transcriptionally active locus • Recombination requires double-strand break at MAT catalyzed by HO endonuclease in a process of gene conversion. Mating-type switching • Cells switch mating type (almost) every generation • Only mother cells (not buds) switch Ash1p is restricted to daughter cells due to mRNA localisation Myosin fibrils locate Ash mRNA to the bud Transcriptional silencing How are the HML and HMRa loci kept transcriptionally silent? • Chromatin structure of HML and HMRa and the surrounding region is condensed (heterochromatin) • Histones are deacetylated • Blocks transcriptional machinery access