* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download talk
Comparative genomic hybridization wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Maurice Wilkins wikipedia , lookup
DNA sequencing wikipedia , lookup
Whole genome sequencing wikipedia , lookup
DNA barcoding wikipedia , lookup
Gel electrophoresis of nucleic acids wikipedia , lookup
Genome evolution wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
DNA supercoil wikipedia , lookup
DNA vaccination wikipedia , lookup
Molecular cloning wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
Point mutation wikipedia , lookup
Bisulfite sequencing wikipedia , lookup
Chloroplast DNA wikipedia , lookup
Light-dependent reactions wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Genomic library wikipedia , lookup
Community fingerprinting wikipedia , lookup
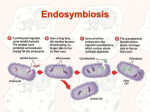
The Origin of Chloroplasts Comparative Analysis of the Genomes of Cyanobacteria and Plants n 1883 – Schimper – Independent binary fission of chloroplasts, and a resemblance of chloroplasts to cyanobacteria Naoki Sato Presented by Ken Ober Akin to Kin? The Origin of Chloroplasts n n n n 1883 – Schimper – Independent binary fission of chloroplasts, and a resemblance of chloroplasts to cyanobacteria 1905 - Mereschkowsky – Endosymbiosis http://www.bio.mtu.edu/~jkoyadom/algae_webpage/charophyceans/Spirogyra_jason4_chassel2_20125.jpg http://tolweb.org/tree/eubacteria/cyanobacteria/prochloron.200.jpeg 1 The Proposed Story The Origin of Chloroplasts n n n www.lifesci.utexas.edu/faculty/sjasper/images/john4.16.jpg http://www.biol.tsukuba.ac.jp/~inouye/ino/gl/Cyanophora.GIF The Origin of Chloroplasts n n n n 1883 – Schimper – Independent binary fission of chloroplasts, and a resemblance of chloroplasts to cyanobacteria 1905 - Mereschkowsky – Endosymbiosis 1924 - Korshikov - Cyanophora paradoxa fossil described – transition organism 1959 - Stocking and Gifford - Independent DNA in chloroplasts 1883 – Schimper – Independent binary fission of chloroplasts, and a resemblance of chloroplasts to cyanobacteria 1905 - Mereschkowsky – Endosymbiosis 1924 - Korshikov - Cyanophora paradoxa fossil described – transition organism The Origin of Chloroplasts n n n n n 1883 – Schimper – Independent binary fission of chloroplasts, and a resemblance of chloroplasts to cyanobacteria 1905 - Mereschkowsky – Endosymbiosis 1924 - Korshikov - Cyanophora paradoxa fossil described – transition organism 1959 - Stocking and Gifford - Independent DNA in chloroplasts 1970 – Margulis – Endosymbiosis PR 2 The Origin of Chloroplasts n n n n n n 1883 – Schimper – Independent binary fission of chloroplasts, and a resemblance of chloroplasts to cyanobacteria 1905 - Mereschkowsky – Endosymbiosis 1924 - Korshikov - Cyanophora paradoxa fossil described – transition organism 1959 - Stocking and Gifford - Independent DNA in chloroplasts 1970 – Margulis – Endosymbiosis PR 1972 - Pigott and Carr - Quantitative relatedness study of Euglena chloroplast DNA and cyanobacterial DNA. The Origin of Chloroplasts n n n n n n n The Origin of Chloroplasts n n n n n n n n 1883 – Schimper – Independent binary fission of chloroplasts, and a resemblance of chloroplasts to cyanobacteria 1905 - Mereschkowsky – Endosymbiosis 1924 - Korshikov - Cyanophora paradoxa fossil described – transition organism 1959 - Stocking and Gifford - Independent DNA in chloroplasts 1970 – Margulis – Endosymbiosis PR 1972 - Pigott and Carr - Quantitative relatedness study of Euglena chloroplast DNA and cyanobacterial DNA. 1978 - Schwartz and Dayhoff – Protein and RNA or DNA sequencing suggests chloroplasts are reduced forms of cyanobacteria 1993 - Hallick et al., Reith and Munholland - Entire chloroplast sequences for Euglena and Porphyra; similarity in organization of genes in operons 1883 – Schimper – Independent binary fission of chloroplasts, and a resemblance of chloroplasts to cyanobacteria 1905 - Mereschkowsky – Endosymbiosis 1924 - Korshikov - Cyanophora paradoxa fossil described – transition organism 1959 - Stocking and Gifford - Independent DNA in chloroplasts 1970 – Margulis – Endosymbiosis PR 1972 - Pigott and Carr - Quantitative relatedness study of Euglena chloroplast DNA and cyanobacterial DNA. 1978 - Schwartz and Dayhoff – Protein and RNA or DNA sequencing suggests chloroplasts are reduced forms of cyanobacteria The Origin of Chloroplasts n n n n n n n n n 1883 – Schimper – Independent binary fission of chloroplasts, and a resemblance of chloroplasts to cyanobacteria 1905 - Mereschkowsky – Endosymbiosis 1924 - Korshikov - Cyanophora paradoxa fossil described – transition organism 1959 - Stocking and Gifford - Independent DNA in chloroplasts 1970 – Margulis – Endosymbiosis PR 1972 - Pigott and Carr - Quantitative relatedness study of Euglena chloroplast DNA and cyanobacterial DNA. 1978 - Schwartz and Dayhoff – Protein and RNA or DNA sequencing suggests chloroplasts are reduced forms of cyanobacteria 1993 - Hallick et al., Reith and Munholland - Entire chloroplast sequences for Euglena And Porphyra; similarity in organization of genes in operons 2002 – Sato – Elucidation of homology in cyanobacteria and plants, using complete genome/proteome sequences 3 Doing It The “Sato” Way! n n The question: What novel information is provided by a whole-genome/proteome comparison of cyanobacteria and plants? Experimental Design: – Look for homology in three species of cyanobacteria – See how the homologies change by adding in three more – Then perform this analysis including a plant species and negative control species The Analysis n Genome Sequences For Your Reference (and Mine) n Cyanobacteria – – – – – – Sy = Synechocystis An = Anabaena Np = Nostoc Pm1 = Prochlorococcus marinus MED4 Pm2 = P. marinus MIT9313 S81 = Synechococcus n Plant n Negative Controls – Ath = Arabidopsis – – – – n Ec = Escherichia Bs = Bacillus Hp = Helicobacter Sc = Saccharomyces Draft Sequences – Rp = Rhodopseudomonas – Rs = Rhodobacter The Results – Short Sequence Features – GC Skew (siseq) – Palindromes (siseq) – Rare Restriction Endonuclease Sites – Dinucleotide Relative Abundance (dinucf) § Phylogenetic tree construction by taking the mean of the differences of DRA’s (phylip) n Proteome sequences – Homology Groups (blast) § Phylogenetic tree construction by the Parsimony method (phylip) 4 The Analysis n Genome Sequences The Results – Dinucleotide Relative Abundances – GC Skew (siseq) – Palindromes (siseq) – Rare Restriction Endonuclease Sites – Dinucleotide Relative Abundance (dinucf) § Phylogenetic tree construction by taking the mean of the differences of DRA’s (phylip) n Proteome sequences – Homology Groups (blast) § Phylogenetic tree construction by the Parsimony method (phylip) The Analysis n Genome The Results – Homology Sequences – GC Skew (siseq) – Palindromes (siseq) – Rare Restriction Endonuclease Sites – Dinucleotide Relative Abundance (dinucf) § Phylogenetic tree construction by taking the mean of the differences of DRA’s (phylip) n Proteome sequences – Homology Groups (blast) § Phylogenetic tree construction by the Parsimony method (phylip) 5 The Results - Homology The Analysis n Genome Sequences – GC Skew (siseq) – Palindromes (siseq) – Rare Restriction Endonuclease Sites – Dinucleotide Relative Abundance (dinucf) § Phylogenetic tree construction by taking the mean of the differences of DRA’s (phylip) n Proteome sequences – Homology Groups (blast) § Phylogenetic tree construction by the Parsimony method (phylip) How do these data answer the original question? The Results - Homology n What novel information is provided by a whole-genome/proteome comparison of cyanobacteria and plants? Higher confidence n More detail n – Bigger data sets – Whole-sequence patterns vs. single gene/protein patterns – Life “core” (238 groups) – Endosymbiosis “core” (80 groups) 6 Acknowledgements n Judith Klein-Seetharaman n Natalie & Scottie ¿Preguntas? Biological Language Modeling Cyanobacteria Photosynthetic Organism 1 Photosynthetic Organism 2 7