* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download TRYPSIN / LYS
Protein domain wikipedia , lookup
Protein folding wikipedia , lookup
Circular dichroism wikipedia , lookup
List of types of proteins wikipedia , lookup
Protein structure prediction wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Alpha helix wikipedia , lookup
Protein purification wikipedia , lookup
Western blot wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
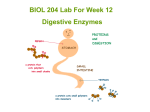
LYS-C + TRYPSIN DIGESTION TCA-precipitated Proteins Add 30µl of 100mM Tris-HCl, pH 8.5, 8M Urea (made fresh). Vortex Bring solution to 5mM TCEP with 0.1M stock @ RT, 30 min Bring solution to 10mM CAM with 0.5M stock @ RT, 30min, in dark Add 1µl Endoproteinase Lys-C @ 1µg/µl @ 37°C for >6 hours Dilute to 2M Urea with 100mM Tris-HCl, pH 8.5 Add CaCl2 to 2mM (stock at 500mM) Add 5µl Trypsin at 0.1µg/µl @ 37°C O/N Add 90% Formic acid to 5% Sample Tris-Urea TCEP CAM Lys-C Tris CaCl2 Ti Formic 30µl 1.5µl 0.6µl 1µl 90µl 0.48µl 5µl 7µl Lys-C EC 3.4.21.50 Inhibitors: DFP, TLCK, aprotinin, and leupeptin Molecular weight: Mr = 33 kD (reduced), Mr = 30 kD (non-reduced) Optimum pH: 8.5 - 8.8 Specificity: Serine protease that specifically hydrolyzes amide, ester, and peptide bonds at the carboxylic side of Lys. Stability: stable in 4 M urea Trypsin EC 3.4.21.4 Inhibitors: TLCK, DFP, PMSF, leupeptin, soybean trypsin inhibitor, trypsin inhibitor from hen egg, aprotinin, α2macroglobulin, α1-antitrypsin, APMSF, and antipain Specificity: Serine endopeptidase that specifically hydrolyzes proteins and peptides at the carboxy side of the basic amino acids Arg and Lys. Amide and ester bonds of Arg and Lys are also cleaved. Optimum pH: 8.0 Description Trypsin specifically hydrolyzes peptide bonds at the carboxylic sides of lysine and arginine residues. Unmodified trypsin is subject to autolysis, generating fragments that can interfere with protein sequencing, HPLC or mass spectrometry analysis of the peptides. In addition, autolysis can result in the generation of pseudotrypsin, which has been shown to exhibit an additional chymotrypsin-like specificity (1). Promega Trypsin has been modified by reductive methylation, rendering it extremely resistant to autolytic digestion (2). In functional stability tests, modified trypsin retains at least two times as much activity as unmodified trypsin after a 3-hour incubation at 37°C. The sequencing grade of modified trypsin has been further improved by TPCK treatment followed by affinity purification yielding a highly active and stable molecule. Sequencing Grade Modified Trypsin is provided in convenient 20μg aliquots with a stability-optimized resuspension buffer. A protease:protein ratio of 1:100 to 1:20 (w/w) is recommended. Recommended Reaction Buffer: 50mM NH4HCO3 (pH 7.8). 1