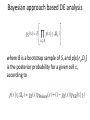* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Bayesian approach to single-cell differential expression analysis
Genomic imprinting wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Designer baby wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Epigenetics in stem-cell differentiation wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Gene expression programming wikipedia , lookup
SCDE: Bayesian approach to singlecell differential expression analysis Presenter: Zheng “Alex” Fu, Ph.D. LIAI, Bioinformatics Core Kharchenko, P. V., Silberstein, L. & Scadden, D. T. Bayesian approach to single-cell differential expression analysis. Nat. Meth. 11, 740-742 Bottleneck in single-cell RNA-Seq analysis Strategy • Datasets – a 92-cell set consisting of mouse embryonic fibroblast (MEF) and embryonic stem (ES) cells. – a data set of cells from different stages of early mouse embryos • Fitting individual error models • Using Bayesian approach to carry out differential expression analysis. Pair-wise comparison of MEF cells. Fitting an individual error model • To fit an individual error model Ωc for a measurement of a single cell c, the observed RPM values were modeled as a function of an expected expression magnitude, with the set of estimates for a subset of genes described previously. Fitting an individual error model • The RPM level rc observed for a gene in cell c was modeled as a mixture of a dropout and amplified components, as a function of an expected expression magnitude e, as • For each cell, the model Ωc was fit with an EM algorithm based on the set of genes for which expected expression magnitudes have been obtained. Fitting an individual error model Bayesian approach based DE analysis With a Bayesian approach, the posterior probability of a gene being expressed at an average level x in a subpopulation of cells S was determined as an expected value (E) according to Bayesian approach based DE analysis where B is a bootstrap sample of S, and p(x|rc,Ωc) is the posterior probability for a given cell c, according to Bayesian approach based DE analysis where pd is the probability of observing a dropout event in cell c for a gene expressed at an average level x in S, pPoisson(x) and pNB(x|rc) are the probabilities of observing expression magnitude of rc in case of a dropout (Poisson) or successful amplification (NB) of a gene expressed at level x in cell c, with the parameters of the distributions determined by the Ωc fit. Bayesian approach based DE analysis For the differential expression analysis, the posterior probability that the gene shows a fold expression difference of f between subpopulations S and G was evaluated as where x is the valid range of expression levels. The posterior distributions were renormalized to unity, and an empirical P value was determined to test for significance of expression difference. Bayesian approach based DE analysis Thanks!