* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download ppt
Cryobiology wikipedia , lookup
Evolution of metal ions in biological systems wikipedia , lookup
Adenosine triphosphate wikipedia , lookup
Citric acid cycle wikipedia , lookup
Fatty acid metabolism wikipedia , lookup
Point mutation wikipedia , lookup
Phosphorylation wikipedia , lookup
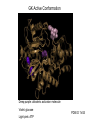
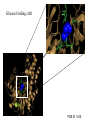
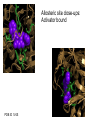
Glucokinase, a glucose sensor Allison Craney November 30, 2006 Glucokinase (GK): Biological Significance • Dramatic conformational change when activated • Allosteric activator binding • Monomeric protein which displays cooperativity • Potential anti-diabetic therapeutic • Current. Crystal structures solved in 2004 GK: basic properties • Catalyzes glucose-6-phosphate production ATP GK • Hexokinase family member with important distinctions – low affinity for glucose – not regulated by end products • Responsible for 20% total catalysis of glucose • 80% in Beta cells (pancreas) and hepatocytes (liver) Molecules: www.steve.gb.com/science/core_metabolism.html Glucose homeostasis is tightly regulated • Mammalian blood glucose level (G): 5mM G: 7-8mM G: <5mM Pancreatic beta cells produce insulin Glucose Uptake Liver alpha cells produce glucagon Stimulates Catabolism GK is a glucose sensor in Beta cells • Triggers insulin release via metabolic coupling of the Na+ and Ca++ channels • Supply-driven pathway – Low affinity for G 1. G blood level INCREASE 2. ADP increases GK activity + increased ATP ATP dependent Na+ channel closes 3. Closing allows the opening of Ca2+ channel, coating cytoplasm with Ca2+ 4. Insulin is released • Activity lowers blood G level Structure Determination •Crystallization via hanging drop method •Structure solved by molecular replacement, using AMORE and CCP4 programs 1V4S 1V4T Resolution (A) 2.3 3.4 Rworking (%) 23.2 23.8 Rfree (%) 27.4 30.6 Average (A)2 B-factor ~40 39.2 Determinants Structural properties of GK •50 kDa, 465 amino acids, mainly alpha helical •Smallest hexokinase –Structurally diverse, but has conserved actin fold QuickTime™ and a Sorenson Video 3 decompressor are needed to see this picture. •Monomer, palm-shaped •Binds G, ATP, activator 1V4S. GK, closed conformation GK Active Conformation Deep purple: allosteric activaton molecule Violet: glucose Light pink: ATP PDB ID 1V4S Glucose binding cleft PDB ID 1V4S Allosteric site close-ups: Activator bound PDB ID 1V4S GK undergoes a large conformational change • Two conformations of GK – CLOSED (active form). Glucose plus allosteric activator bound. – OPEN (inactive form). No glucose bound. • Between two states, huge rotation of 120 degrees. • Conversion is much slower than catalytic activity – Only monomeric protein to display cooperativity – Hill coefficient = 1.9 Deep blue: flexible loop which covers G binding Green: Small subunit G binding residues Teal: Asp205 (large subunit) Purple: Activator binding residues in allosteric site PDB ID 1V4T GK Conformational Change Closed conformation PDB ID 1V4S “Super-open” conformation PDB ID 1V4T GK mutations decrease glucose homeostasis • >200 human GK mutations have been isolated • (25-30% glycolysis in Beta cells: 5mM blood G) • Loss-of-function mutations: much more G needed to reach glycolytic activity level. Elevated blood G. Condition: early onset diabetes mellitus • Gain-of-function mutations: less G is needed. Condition: hypoglycemia. Patients’ blood G: 1.5-3mM. • Potential antidiabetic drug- screen of >120,000 compounds. RO-28-1675 decreased blood G while increasing insulin levels above basal. QuickTime™ and a Sorenson Video 3 decompressor are needed to see this picture. Selected Literature • • • • • Grimsby, et al. 2003. Allosteric activators of Glucokinase: potential role in diabetes therapy. Science. 301: 370-373. Kamata, et al. 2004. Structural basis for allosteric regulation of the monomeric allosteric enzyme human glucokinase. Structure. 12: 429-438. Zelent, et al. 2005. Glucokinase and glucose homeostasis: proven concepts and new ideas. Bioch. Soc. Trans. 33 (1): 306-310. PDB ID 1V4S and 1V4T (Kamata, et al., 2004) were morphed using Yale Morph Server All movies and figures were produced in Pymol Molecular Graphics System (DeLano Scientific LLC, San Carlos, CA; http://www.pymol.org).