* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download protein - Portal UniMAP
Signal transduction wikipedia , lookup
Paracrine signalling wikipedia , lookup
Peptide synthesis wikipedia , lookup
Gene expression wikipedia , lookup
Expression vector wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Magnesium transporter wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Point mutation wikipedia , lookup
Genetic code wikipedia , lookup
Interactome wikipedia , lookup
Protein purification wikipedia , lookup
Biosynthesis wikipedia , lookup
Homology modeling wikipedia , lookup
Western blot wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Metalloprotein wikipedia , lookup
Protein–protein interaction wikipedia , lookup
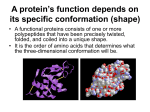
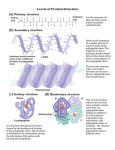
ERT106 BIOCHEMISTRY PROTEIN Pn Syazni Zainul Kamal Protein structure Protein (polypeptides) are organic compound made of amino acids arranged in a linear form and folded into specific conformation Protein - essential part of organisms - participate virtually in every process within cells Protein structure When cell synthesizes a polypeptides, the chain folded spontaneously This folding is reinforced by variety of bonds between the chain In a complex structure of protein, several levels of the structural organization of proteins : a) primary structure c) tertiary structure b) secondary structure d) quaternary structure a) Primary structure Primary structure of protein is its unique sequence of amino acids forming its polypeptide chain Every polypeptides has a specific amino acid sequence the primary structure of a protein is starting from the amino-terminal (N) end to the carboxyl-terminal (C) end Primary structure of enzyme lysozyme b) Secondary structure Most proteins have segments of their polypeptide chain repeatedly coiled of folded in patterns. These coiled & folded referred as secondary structure. 2 types of secondary structure : - α-helix stabilized by hydrogen bond - β-pleated sheet between carbonyl & amino groups in the polypeptide’s backbone α-helix Rigid, rodlike structure when a polypeptides chain twists into a right-handed helical conformation Hydrogen bond form between amino group of each amino acid and the carbonyl group of the amino acid four residue away (H bond form between 4 amino acid) R group extend outward from the helix β-pleated sheet Form when two or more polypeptide chain segments line up side by side Each individual segment = β-strand Each β-strand is fully extended β-pleated sheet stabilized by hydrogen bonds form between the polypeptide backbone N-H and carbonyl groups of adjacent chains Two β-pleated sheet : - parallel - polypeptide chain arranged in same direction - antiparallel - polypeptide chain arranged in opposite direction - more stable Usually mixed parellal-antiparallel β-pleated sheet observed in proteins combination of α-helix and β-pleated sheet secondary structure = supersecondary structure Supersecondary structure : a) βαβ unit two parallel β-pleated sheets connected by α-helix fragment b) β-meander two antiparallel β-sheets are connected by polar amino acids and glycines to effect an abrupt change in direction of the polypeptide chain (reverse or β-turns) b meander c) αα-units two α-helices separated by loop or nonhelical segment d) β-barrel form when various β-sheet configurations fold back on themselves e) Greek key antiparallel β-sheet doubles back on itself in a pattern that resemble a common greek pottery design c) Tertiary structure Three-dimensional structure of a single protein molecules The α-helices and β-pleated sheets are folded into compact globule. Protein folding occurs as consequence of interactions between the side chains in their primary structure Interactions that stabilize tertiary structure 1) Hydrophobic interactions As polypeptide folds, amino acids with hydrophobic (nonpolar) side chain are brought close to each other, out of contact with water. 2) Electrostatic interactions Interaction occurs between ionic groups of opposite charge (referred as salt bridge) 3) Hydrogen bonds Large number of hydrogen bond form within a protein’s interior and on its surface Examples of amino acid side chains that may hydrogen bond to each other: Two alcohols: ser, thr, and tyr. Alcohol and an acid: asp and tyr Two acids: asp and glu Alcohol and amine: ser and lys Alcohol and amide: ser and asn 4) Covalent bond Created by chemical reactions that alter a polypeptide's structure during or after its synthesis eg. Disulphide bond (strong linkage) Protect protein structure from adverse changes in pH or salt concentrations d) Quaternary structure Proteins consist of two or more polypeptide chains aggregated into one functional macromolecules Many proteins, esp those with high molecular weight are composed of several polypeptide chains. In proteins that consist of more than 1 polypeptide chain, each polypeptide is called subunit Polypeptide subunits assemble and held together by noncovalent interaction eg H bonding, hydrophobic effect, electrostatic interaction Loss of protein structure Protein structure is sensitive to environment factors Many physical & chemical agents can disrupt protein’s conformation The process of structure disruption = denaturation Denaturing conditions : Strong acid or base – changes in pH result in protonation of some protein side group, which alter/disrupt hydrogen bonding & salt bridge Organic solvents – water-soluble organic solvents eg. Ethanol interfere with hydrophobic interaction Detergents – these amphiphatic molecules disrupt hydrophobic interaction causing proteins to unfold into extended polypeptide chains (amphiphatic = contain nonpolar and polar components) Reducing agents – eg. Urea, βmercaptoethanol, will convert disulfide bridge (S-S) to sulfhydryl group (SH) urea disrupt H bond & hydrophobic interaction Heavy metal ions – mercury (Hg+) and lead (Pb2+) disrupt salt bridge by forming ionic bond with negatively charge group. Temperature change – as temp increase, the rate of molecular vibration increase. So weak H bond will be disrupt and protein will unfold. Mechanical stress – stirring & grinding actions disrupt the delicate balance of forces that maintain protein strcuture. eg. Foam formed when egg white is beaten vigorously contains denatured protein Fibrous proteins Fibrous protein exist as a long stranded molecules Contain high proportions of secondary structures ; α-helices and β-pleated sheets Most are structural protein eg. α-keratin, collagen, silk fibroin α-keratin Found in hair, wool, skin, horns, fingernails is an α-helical polypeptides Each polypeptide has three domain : - an amino terminal ‘head’ - a central rodlike α-helical domain - a carboxyl terminal ‘tail’ Two keratin polypeptides associate to form = coiled coil dimer Two keratin polypeptides associate to form = coiled coil dimer Two antiparallel rows of these dimer form a supercoiled structure called a protofilament (disulfide bond aid the formation of protofilament) Hundreds of filaments, each containing 4 protofilaments form macrofibril Each hair cells (fiber) contain several macrofibrils Collagen The most abundant protein in vertebrates Synthesized by - connective tissue cells mostly found in fibrous tissues such as : tendon, ligament and skin, in cornea, cartilage, bone, blood vessels, the gut Extremely strong Collagen is composed of three polypeptide helices that are twisted around each other to form a triple helix (stabilized by hydrogen bonding) The amino acid composition of collagen is distinctive - high content of glycine, proline and lysine - very little amount of cysteine (unlike αkeratin) Silk fibroin Silk protein - form spider webs, cocoon and nests - consist of the fibrous protein fibroin Considered to be β-keratin - polypeptide chains arranged in antiparallel βpleated sheet comformation Its primary structure mainly consists of the amino acid sequence (Gly-Ser-Gly-Ala-GlyAla)n Globular protein Globular protein have a spherical shape, compact and water-soluble In their function, usually require them to bind precisely to other molecules Most enzyme are globular Myoglobin & hemoglobin are typical example of globular protein Both are hemoprotein and each is involved in oxygen metabolism Unlike fibrous proteins which only play a structural function, globular proteins can act as: 1) Enzymes, by catalyzing organic reactions taking place in the organism in mild conditions and with a great specificity. 2) Messengers, by transmitting messages to regulate biological processes. This function is done by hormones, i.e. insulin etc. 3) Transporters of other molecules through membranes 4) Stocks of amino acids. 5) Regulatory roles are also performed by globular proteins rather than fibrous proteins. a) Myoglobin Myoglobin is an iron- and oxygen-transport protein found in the muscle tissue of vertebrates in general and in almost all mammals. composed of a single polypeptide chain of 153 amino acid residues and has the ability to store oxygen by binding it to an iron atom (heme) It is found abundantly in the tissues of diving mammals, e.g., the whale, the seal, and the dolphin. High concentrations of myoglobin in these animals, allows them to store sufficient oxygen to remain underwater for long periods. A myoglobin polypeptide is comprised of 8 separate right handed α-helices, that are connected by short non helical regions. Each myoglobin molecule contains one heme prosthetic group Each heme consist of porphyrin ring with Fe2+ in the center Free heme [Fe2+] has a high affinity for O2 and is irreversibly oxidized to form hematin [Fe3+] b) Hemoglobin Hemoglobin is a roughly spherical molecule found in red blood cells Function = transport oxygen from lung to every tissues in the body Composed - two α-chain - two β-chain The protein contain four subunits, designated α and β. Each subunit contain a heme group that bind with oxygen