* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Response of the Innate Immune System to Pathogens
Monoclonal antibody wikipedia , lookup
Plant disease resistance wikipedia , lookup
Complement system wikipedia , lookup
Adoptive cell transfer wikipedia , lookup
Hygiene hypothesis wikipedia , lookup
Drosophila melanogaster wikipedia , lookup
DNA vaccination wikipedia , lookup
Immune system wikipedia , lookup
Cancer immunotherapy wikipedia , lookup
Molecular mimicry wikipedia , lookup
Adaptive immune system wikipedia , lookup
Polyclonal B cell response wikipedia , lookup
Immunosuppressive drug wikipedia , lookup
Psychoneuroimmunology wikipedia , lookup
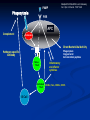
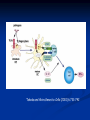
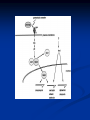
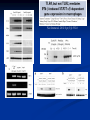
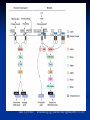
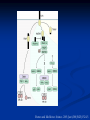
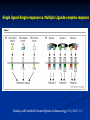
Response of the Innate Immune System to Pathogens: Pattern Recognition Receptors What is the Innate Immune Response? A universal and evolutionarily conserved mechanism of host defense against infection First line of Defense Predates the adaptive immune response Found in all multicellular organisms Adaptive only in vertebrates Uses receptors and effectors that are ancient in their lineage Must provide protection against a wide variety of pathogens Distinguishes self from non-self perfectly Defects in innate immunity are very rare and almost always lethal The Innate Immune Response: Common Misconceptions The innate immune system is an evolutionary rudiment whose only function is to contain the infection until the “real” immune response can kick in. Adaptive immunity developed because of the inflexibility of the nonclonal receptors used by the innate immune response. The innate system cannot cope with the high mutational rate and heterogeneity of pathogenic organisms. The Innate immune system instructs the adaptive immune response to respond to microbial infection The major decision to respond or not respond to a particular ligand is decided by the genome-encoded receptors of the innate immune system Adapted from Medzhitov and Janeway Cur. Opin. Immunol. 1997 9:4-9 PAMP Phagocytosis Y PRR APC Complement Endosome MHC Pathogen-specific Antibody Direct Bactericidal Activity Phagocytosis Oxygen burst Anti-microbial peptides B7 Y Naive T Cell Inflammatory and effector cytokines Activated CD40L, FasL, CD30L, CD27L T Cell B Cell Components of Innate and Adaptive Immunity Innate Immunity Adaptive Immunity Physical Barriers skin, gut villi, lung cilia,etc none Soluble Factors many protein and non-protein secretions Immunoglobulins (antibody) Cells phagocytes, NK cell eosinophils T and B lymphocytes PAMPs: Pathogen Associated Molecular Patterns PRRs: Pattern Recognition Receptors Takeda and Akira Genes to Cells (2001) 6:733-742 The NF-kB Family of Transcription Factors Eukaryotic transcription factor found in essentially all cell types First described in 1986 as a nuclear factor required for the transcription of the immunoglobulin kappa light chain in B cells. Binds to a 10-bp sequence GGGGYNNCCY Important component in the inducible expression of many proteins: cytokines, acute phase proteins, adhesion molecules The NF-kB signaling system is evolutionarily conserved Three NF-kB molecules in Drosophila dorsal controls dorsal/ventral polarity during development Regulates antifungal gene expression dif and relish: regulate expression of antifungal and antibacterial genes NF-kB exists in the cytoplasm as an inactive heterotrimer composed of 2 Rel family proteins and an inhibitory IkB molecule Stress, infection, or cytokine IKK P P (Ub)n IkB p65 p50 26S proteosome Nuclear Translocation Activation of NF-KB Responsive genes NF-kB Family Structure Conservation of signaling pathways between flies and humans Toll Pelle tube ? ? cactus kinase cactus dorsal dif relish Mutation of genes in the Toll signaling cascade in Drosophila block the expression of anti-fungal and anti-bacterial genes Water A. fumigatus E. coli Signaling to monocytic cells through the human Toll homolog induces accessory cell functions NF-kB Activation Requirements for the recognition of targets by the innate immune Molecular structures recognized by the immune system must be shared by large groups of pathogens molecular patterns vs. particular structures (antigens) PAMP PAMPs must be conserved products of microbial metabolism not subject to antigenic variability The recognized structures must be absolutely distinct from self antigens: discrimination of self vs. non-self Molecules involved in the recognition of LPS by Cells LPS Binding Protein (LBP) 60-kDa serum glycoprotein that binds with high affinity to LPS Sequence homology to bactericidal/permeability increasing protein Acute phase protein secreted by hepatocytes Serum levels between 1 and 10 ug/ml in normal human serum Concentrations >300 ug/ml in acute phase serum Critical for rapid responses to small amounts of LPS or gramnegative bacteria and for survival of Salmonella infection Expression of LBP in hepatocytes is regulated by LPS, IL-1, IL-6 and TNF Molecules involved in the recognition of LPS by Cells CD14, sCD14 CD11b/CD18 (Mac1) 55 kDa GPI-linked protein on the surface on monocytes and PMN Also found as a soluble protein in serum:sCD14 Each CD14 molecule binds 1 molecule of LPS complexed to LBP Required for responses of genes to low concentrations of LPS No cellular signaling activity Alternative cell surface receptor for LPS Recognizes LPS at high concentrations For expression of a full repertoire of LPS-inducible genes, CD14 and CD11b/CD18 must be coordinately engaged to deliver optimal signaling to the macrophage. Search for the LPS Gene Endotoxin hyporesponsive mice C3H/HeN: Normal LPS response 1. C3H/He 1947 Spontaneous mutation mid 1960’s 2. C57BL/10Sn Sometime After 1953 C3H/HeJ: LPS hyporesponsive C57BL10/ScCR LPS hyporesponsive January 1998 DNAX reports the cloning of human TLR1-5 No ligands described Janeway’s hToll=TLR4 Rock, et al. 1998 PNAS 95:588 September 1998 Genentech identifies TLR2 as the LPS signaling molecule by transfection studies Didn’t look at TLR4 December 1998 Tularik also identifies TLR2 as LPS signaling molecule in transfection studies TLR4 didn’t work September 1998: Bruce Buetler’s lab mapped LPS to a 1.2Mb region of chromosome 4. The only intact gene within this region is TLR4 December 1998: “Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: Mutations in Tlr4 gene” Poltarak et al. Science 282:2085 Poltarak et al 1998 Science 282:2085 A single nucleotide change in the gene for TLR4 renders the C3H/HeJ mouse non-responsive to LPS C3HHeN RFHLCLHYRDFI P GCAIAANIIQEGFHK C3HHeJ RFHLCLHYRDFI H GCAIAANIIQEGFHK Hoshino et al 1999 J. Immunol. 162:3749 TLR4 knockout mice are hyporesponsive to LPS Dunne and O’Neil 2003 www.stke.org/cgi/content/full/sigtrans;2003/171/re3 Receptor Agonist(s) (Pattern Recognition Receptors) (Pathogen-Associated Molecular Patterns) TLR1 Heterodimerizes with TLR2 TLR2 PGN, some LPS, some LTA, lipoproteins, AraLAM TLR3 dsRNA TLR4 Gram(-) LPS, Taxol, some LTA TLR5 Flagellin TLR6 Heterodimerizes with TLR2 TLR7 Imidazoquinoline TLR9 Bacterial DNA (CpG) TLR 8,10 Unknown Akira J.Biol.Chem. 2003 278:38105–38108 How do we explain the Genentech and Tularik findings? How could TLR4 knockout mice be LPS-nonresponsive but TLR2 transfected cells LPS responsive? Why were TLR4-transfected cells LPS non-reponsive? 9.0 8.0 In both cases, the LPS preparations used by the investigators were contaminated with bacterial lipopeptides: TLR2 agonists! 7.0 Fold Induction 1. 6.0 5.0 4.0 3.0 2.0 1.0 0.0 TLR2 Control TLR2 Sigma LPS TLR2 pure LPS TLR2 LTA TLR2 PGN TLR4 Control TLR4 Sigma LPS TLR4 pure LPS 0.6000 0.5000 Vector 2. In Tularik’s experiments with transfected TLR4, they lacked a critical accessory protein required for efficient TLR4 expression and function: MD-2 Relative Light Units MD2 0.4000 0.3000 0.2000 0.1000 0.0000 Control LPS PMA TLR4 LTA TLR4 PGN Common Themes in IL-1, TLR, and IL-18 Signaling TLR4/4 IL-1RI IL-1RAcP CD14 MD-2 MyD88 MyD88 IRAK IL-18R MyD88 IRAK TRAF6 IL-1RAcP IRAK TAK1/ NIK IKK Complex IkB p65 p50 Death domain TIR domain NF-kB activation Nat Immunol. 2002 Apr;3(4):392-8 Nature 2003 420:329 TIRAP=MAL Nature 2003 420:324 Luke A.J. O’Neil www.stke.org/cgi/content/full/sigtrans;2003/171/re3 Science. 2003 Aug 1;301(5633):640-3 Barton and Medzhitov Science. 2003 Jun 6;300(5625):1524-5 Common and Distinct Themes in TLR Signaling TLR4/4 TLR2 CD14 TLR1/6 Rac MD-2 PI3K TRIF PI3K TIRAP MyD88 MyD88 IRAK IRF3 TIRAP IRAK TRAF6 TAK1/ NIK AKT MAP kinases IFN-b Death domain IKK Complex TIR domain IkB p65 p50 TLR2Specific Common Responses TLR4Specific Single ligand-Single response vs. Multiple Ligands-complex response Ozinsky and Underhill Current Opinion in Immunology 2002, 14:103–110 Other PRRs Dectin-1 Macrophage Mannose Receptor C-type lectin that binds b-glucan-containing particles including zymosan and C. albicans Cell bound C type lectin that binds sugar molecules on the surface on many bacteria and viruses. Macrophage Scavenger Receptor (SR-A) Recognize certain anionic polymers and low-density lipoproteins 4 forms in humans: Peptidoglycan Recognition Proteins (PGRP) PGRP-L: liver PGRP-Ia and PGRP-Ib: esophagus PGRP-S: bone marrow Adjuvants Functionally defined as any substance that, when mixed with antigen, increases the immunogenicity of the antigen Injection of unmodified, nonself proteins in the absence of adjuvant results in tolerance.