Gene expression
... • nucleotide sequence encoded by a gene that remains present within the final mature RNA product of that gene after introns have been removed by RNA splicing. • This is the expressed genetic material… the light is turned on. ...
... • nucleotide sequence encoded by a gene that remains present within the final mature RNA product of that gene after introns have been removed by RNA splicing. • This is the expressed genetic material… the light is turned on. ...
DNA-binding motifs
... or histone proteins is associated with the control of gene expression. • Clusters of methylated cytosine nucleotides bind to a protein that prevents activators from binding to DNA. • Methylated histone proteins are associated with inactive regions of chromatin. ...
... or histone proteins is associated with the control of gene expression. • Clusters of methylated cytosine nucleotides bind to a protein that prevents activators from binding to DNA. • Methylated histone proteins are associated with inactive regions of chromatin. ...
Control of Gene Expression
... or histone proteins is associated with the control of gene expression. • Clusters of methylated cytosine nucleotides bind to a protein that prevents activators from binding to DNA. • Methylated histone proteins are associated with inactive regions of chromatin. ...
... or histone proteins is associated with the control of gene expression. • Clusters of methylated cytosine nucleotides bind to a protein that prevents activators from binding to DNA. • Methylated histone proteins are associated with inactive regions of chromatin. ...
Biology 340 Molecular Biology
... 3. Transcription factors a. DNA binding domains b. activation domains 4. RNA polymerase II 5. Steroid hormone regulation Lecture: 1. Eukaryotic transcription Eukaryotes: --Most are multicellular and made of different cell types. --Different cells express distinct subsets of genes. --Gene expression ...
... 3. Transcription factors a. DNA binding domains b. activation domains 4. RNA polymerase II 5. Steroid hormone regulation Lecture: 1. Eukaryotic transcription Eukaryotes: --Most are multicellular and made of different cell types. --Different cells express distinct subsets of genes. --Gene expression ...
Gene Expression
... • Enhancer are noncoding control sequences that produce transcription, this must be activated for its associated gene to be expressed. • Transcription factors bind to enhancers and RNA polymerase to regulate transcription • Many enhancers are located far away from their genes they need to activate. ...
... • Enhancer are noncoding control sequences that produce transcription, this must be activated for its associated gene to be expressed. • Transcription factors bind to enhancers and RNA polymerase to regulate transcription • Many enhancers are located far away from their genes they need to activate. ...
Lac operon - positive regulation Gene expression of eukaryotic cells
... paternal allele of a gene in early development • certain genes are expressed in a parent-of-originspecific manner Epigenetic regulation ...
... paternal allele of a gene in early development • certain genes are expressed in a parent-of-originspecific manner Epigenetic regulation ...
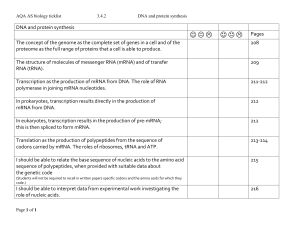
doc 3.4.2 protein synthesis checklist
... Transcription as the production of mRNA from DNA. The role of RNA polymerase in joining mRNA nucleotides. ...
... Transcription as the production of mRNA from DNA. The role of RNA polymerase in joining mRNA nucleotides. ...
regulatory transcription factors
... chromosomes during interphase – During gene activation, tightly packed chromatin must be converted to an open conformation in order for transcription to occur ...
... chromosomes during interphase – During gene activation, tightly packed chromatin must be converted to an open conformation in order for transcription to occur ...
Recent progress in understanding transcription factor binding
... Recent progress in understanding transcription factor binding specificity Gene expression levels can vary greatly from gene to gene and between individuals. To understand how these differences arise, and be able to predict and manipulate them, we need to dissect the molecular mechanisms by which the ...
... Recent progress in understanding transcription factor binding specificity Gene expression levels can vary greatly from gene to gene and between individuals. To understand how these differences arise, and be able to predict and manipulate them, we need to dissect the molecular mechanisms by which the ...
Popular scientific report
... are in the same flower and represent the female and male reproductive tissue respectively, this is called a hermaphroditic flower. The pollination needs a medium, for example, wind or insects. The insect pollination was an evolutionary success, because less pollen is produced by the plant and the fe ...
... are in the same flower and represent the female and male reproductive tissue respectively, this is called a hermaphroditic flower. The pollination needs a medium, for example, wind or insects. The insect pollination was an evolutionary success, because less pollen is produced by the plant and the fe ...
What are transcription factors?
... transcription factor. Transcription factors are proteins with a specific job: they bind the regulatory/non-coding DNA of a gene which will then cause the gene (coding DNA) to be expressed (transcribe into RNA). If a transcription factor is not present, then theoretically nothing will bind that DNA ...
... transcription factor. Transcription factors are proteins with a specific job: they bind the regulatory/non-coding DNA of a gene which will then cause the gene (coding DNA) to be expressed (transcribe into RNA). If a transcription factor is not present, then theoretically nothing will bind that DNA ...
PERSISTENCE: Mechanisms underlying the “Central Dogma
... E. mature mRNA travels out to the cytoplasm where it makes a single protein ...
... E. mature mRNA travels out to the cytoplasm where it makes a single protein ...
Regulation of Gene Transcription
... The TFII complexes are oftern referred to as basal or general transcription factors, are they are required for RNA polymerase II to initiate transcription. ...
... The TFII complexes are oftern referred to as basal or general transcription factors, are they are required for RNA polymerase II to initiate transcription. ...
Gene Section TFEB (transcription factor EB) Atlas of Genetics and Cytogenetics
... Transcription factor; member of the basic-helix-loophelix leucine-zipper transcription factor MiTF/ TFE family (also known as the MiT family), which also contains MiTF, TFEC, and TFE3. The four members form homo- and/or heterodimers to bind the Ebox core sequence CAYGTG; the helix-loop-helix-leucine ...
... Transcription factor; member of the basic-helix-loophelix leucine-zipper transcription factor MiTF/ TFE family (also known as the MiT family), which also contains MiTF, TFEC, and TFE3. The four members form homo- and/or heterodimers to bind the Ebox core sequence CAYGTG; the helix-loop-helix-leucine ...
Recombinant DNA Technology
... to masking by proteins. Fertilization of the egg initiates unmasking and translation of these mRNAs. • Availability of specific tRNAs – In the embryonic development of a hornworm, an mRNA is present from day 1 but a specific tRNA needed for its translation is not produced until day 6. ...
... to masking by proteins. Fertilization of the egg initiates unmasking and translation of these mRNAs. • Availability of specific tRNAs – In the embryonic development of a hornworm, an mRNA is present from day 1 but a specific tRNA needed for its translation is not produced until day 6. ...
Title - Iowa State University
... d. simultaneous polysomes during transcription e. RNA peocessing 18. Combinatorial regulation of gene expression allows: a. regulation of many genes by combinations of a few specific transcription factors. b. regulation of many genes by combinations of large specific transcription factors. c. regula ...
... d. simultaneous polysomes during transcription e. RNA peocessing 18. Combinatorial regulation of gene expression allows: a. regulation of many genes by combinations of a few specific transcription factors. b. regulation of many genes by combinations of large specific transcription factors. c. regula ...
Total Number with GO terms
... Number of genes with this GO term for genes closest to each CNE Number of genes with this GO term for all human genes ...
... Number of genes with this GO term for genes closest to each CNE Number of genes with this GO term for all human genes ...
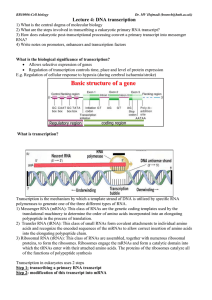
Lecture 4: DNA transcription
... Performed by spliceosomes (large RNA-protein complex made of small nuclear ribonucleoproteins) Recognise exon-intron boundaries and splice exons together by transesterification reactions Cell type-specific splicing ...
... Performed by spliceosomes (large RNA-protein complex made of small nuclear ribonucleoproteins) Recognise exon-intron boundaries and splice exons together by transesterification reactions Cell type-specific splicing ...
Eukaryotic transcriptional control
... The binding of transcription factors to the major groove of DNA •As with most bacterial activators and repressors, a helices in the DNA-binding domain of eukaryotic transcription factors are often oriented so that they lie in the major groove of DNA helix where atoms of protein and DNA make contact ...
... The binding of transcription factors to the major groove of DNA •As with most bacterial activators and repressors, a helices in the DNA-binding domain of eukaryotic transcription factors are often oriented so that they lie in the major groove of DNA helix where atoms of protein and DNA make contact ...
Lecture 40_GeneRegulationI_transcriptional_control_RoadMap
... 1. The Nucleus: Transcription and translation are physically separated in eukaryotes à some regulatory mechanisms, like attenuation, are specific to prokaryotes. 2. Activation vs Repression: Positive regulation may be more predominant in eukaryotes; negative regulation may be more predominant in pr ...
... 1. The Nucleus: Transcription and translation are physically separated in eukaryotes à some regulatory mechanisms, like attenuation, are specific to prokaryotes. 2. Activation vs Repression: Positive regulation may be more predominant in eukaryotes; negative regulation may be more predominant in pr ...
15.2 Regulation of Transcription & Translation
... other hormone mechanism, which involves lipid-soluble hormones (such as oestrogen). Protein Hormones ...
... other hormone mechanism, which involves lipid-soluble hormones (such as oestrogen). Protein Hormones ...
650 BIOLCHEM Fall 2016 Course Announcement
... areas. The course seeks to develop the students’ understanding of recent progress in the investigation of gene expression that is based on advances in biochemical, structural, molecular, cellular and genomic approaches. The lectures are designed to update and to complement survey courses by focusing ...
... areas. The course seeks to develop the students’ understanding of recent progress in the investigation of gene expression that is based on advances in biochemical, structural, molecular, cellular and genomic approaches. The lectures are designed to update and to complement survey courses by focusing ...
第一次课件第八章
... effective when the DNA is joined into a circle by a protein bridge. An enhancer and promoter on separate circular DNAs do not interact, but can interact when the two molecules are catenated. ...
... effective when the DNA is joined into a circle by a protein bridge. An enhancer and promoter on separate circular DNAs do not interact, but can interact when the two molecules are catenated. ...
Transcription factor
In molecular biology and genetics, a transcription factor (sometimes called a sequence-specific DNA-binding factor) is a protein that binds to specific DNA sequences, thereby controlling the rate of transcription of genetic information from DNA to messenger RNA. Transcription factors perform this function alone or with other proteins in a complex, by promoting (as an activator), or blocking (as a repressor) the recruitment of RNA polymerase (the enzyme that performs the transcription of genetic information from DNA to RNA) to specific genes.A defining feature of transcription factors is that they contain one or more DNA-binding domains (DBDs), which attach to specific sequences of DNA adjacent to the genes that they regulate. Additional proteins such as coactivators, chromatin remodelers, histone acetylases, deacetylases, kinases, and methylases, while also playing crucial roles in gene regulation, lack DNA-binding domains, and, therefore, are not classified as transcription factors.