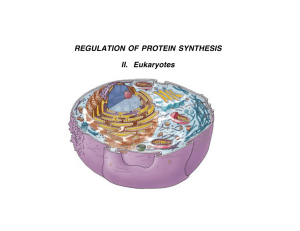
Regulating Protein Synthesis
... generally under positive control (proteins promote, rather than inhibit, RNA polymerase binding to DNA template). ...
... generally under positive control (proteins promote, rather than inhibit, RNA polymerase binding to DNA template). ...
6CDE Transcription and Translation
... helix unzips itself, and the antisense strand of the DNA is transcribed into mRNA. 2. Translation is the process of synthesizing proteins from RNA. The mRNA from transcription carries genetic information from the nucleus to the ribosome for protein synthesis. RNA catalyzes translation and reads the ...
... helix unzips itself, and the antisense strand of the DNA is transcribed into mRNA. 2. Translation is the process of synthesizing proteins from RNA. The mRNA from transcription carries genetic information from the nucleus to the ribosome for protein synthesis. RNA catalyzes translation and reads the ...
Protein Synthesis SG
... 24. What forms can a viral genome take? 25. Describe the lytic and lysogenic infection cycles. Compare & contrast how they allow viruses to spread. 26. Describe the life cycle of a retrovirus. 27. Describe the basic concept of the operon, including the role of each of the following: promoter, regula ...
... 24. What forms can a viral genome take? 25. Describe the lytic and lysogenic infection cycles. Compare & contrast how they allow viruses to spread. 26. Describe the life cycle of a retrovirus. 27. Describe the basic concept of the operon, including the role of each of the following: promoter, regula ...
Chapter 7A
... The control of gene expression by transcription activation and repression has been studied extensively in bacteria. As an example, the E. coli lac operon, which encodes 3 genes (lacZYA) involved in lactose metabolism, uses both mechanisms of control (Fig. 7.3). A specific repressor protein (the lac ...
... The control of gene expression by transcription activation and repression has been studied extensively in bacteria. As an example, the E. coli lac operon, which encodes 3 genes (lacZYA) involved in lactose metabolism, uses both mechanisms of control (Fig. 7.3). A specific repressor protein (the lac ...
File - EUREKA! Science
... Often include sequences that control its expression Two major control sequences are: Promoters Operators ...
... Often include sequences that control its expression Two major control sequences are: Promoters Operators ...
Control of Gene Expression 3 - Dr. Kordula
... I. Transcriptional Control in Eucaryotes – The primary purpose of this kind of control in multicellular organisms is to orchestrate the up and down modulation of specific genes during development and differentiation. Most eukaryotic genes are positively regulated. We will concentrate on Pol II g ...
... I. Transcriptional Control in Eucaryotes – The primary purpose of this kind of control in multicellular organisms is to orchestrate the up and down modulation of specific genes during development and differentiation. Most eukaryotic genes are positively regulated. We will concentrate on Pol II g ...
Recombinant DNA I
... Two types of transcription factors bind enhancers and affect levels of txn: true activators and anti-repressors ...
... Two types of transcription factors bind enhancers and affect levels of txn: true activators and anti-repressors ...
Additional information
... Brief Description of Research: We aim to decipher the complex pathways that control transcription and how cells maintain their transcriptional state via chromatin. These are central basic questions for many biological systems, including cancer and other human diseases. We use yeast as a model organi ...
... Brief Description of Research: We aim to decipher the complex pathways that control transcription and how cells maintain their transcriptional state via chromatin. These are central basic questions for many biological systems, including cancer and other human diseases. We use yeast as a model organi ...
Presentation - people.vcu.edu
... A transcription factor is a protein that binds to specific DNA sequences, thereby controlling the rate of transcription of genetic information from DNA to messenger RNA ...
... A transcription factor is a protein that binds to specific DNA sequences, thereby controlling the rate of transcription of genetic information from DNA to messenger RNA ...
Genomics presentation
... • Nuclear periphery and heterochromatin vicinity areas share a decreased mobility and are genomic silencing regions, while the centre is dynamic and transcriptionally active. • Gene kissing is the intra (cis) or interchromosomal (trans) overlap of sequences within the same position, causing transcri ...
... • Nuclear periphery and heterochromatin vicinity areas share a decreased mobility and are genomic silencing regions, while the centre is dynamic and transcriptionally active. • Gene kissing is the intra (cis) or interchromosomal (trans) overlap of sequences within the same position, causing transcri ...
presentation source
... – Operator is second regulatory site, adjacent to promoter fig 16.12 – lac repressor binds to operator, only when lactose absent – Repressor covers part of promoter when bound to operator ...
... – Operator is second regulatory site, adjacent to promoter fig 16.12 – lac repressor binds to operator, only when lactose absent – Repressor covers part of promoter when bound to operator ...
Test: Gene Regulation Free Response Questions It is known that
... The p53 protein can activate genes that halt the cell cycle by binding to CDKs The proteins could signal pathways that inhibit or halt the cell cycle by binding to miRNAs v. The proteins can activate suicide genes – leading the cell to apoptosis 4. Bacteria often respond to environmental change by r ...
... The p53 protein can activate genes that halt the cell cycle by binding to CDKs The proteins could signal pathways that inhibit or halt the cell cycle by binding to miRNAs v. The proteins can activate suicide genes – leading the cell to apoptosis 4. Bacteria often respond to environmental change by r ...
Slide 1
... • The base pair where transcription initiates is termed the transcription-initiation site or start site • By convention, the transcription-initiation site in the DNA sequence is designated +1, and base pairs extending in the direction of transcription (downstream) are assigned positive numbers which ...
... • The base pair where transcription initiates is termed the transcription-initiation site or start site • By convention, the transcription-initiation site in the DNA sequence is designated +1, and base pairs extending in the direction of transcription (downstream) are assigned positive numbers which ...
BIOL 112 – Principles of Zoology
... 4) Chromatin conformation (remodelling) a. Antirepressors & nucleosome positioning. b. Histone acetylation – (acetyl groups on lysines), histone acetyltransferase enzyme catalyzes the addition of lysine, targeted to genes by specific TFs. c. Heterochromatin – highly condensed, transcriptionally iner ...
... 4) Chromatin conformation (remodelling) a. Antirepressors & nucleosome positioning. b. Histone acetylation – (acetyl groups on lysines), histone acetyltransferase enzyme catalyzes the addition of lysine, targeted to genes by specific TFs. c. Heterochromatin – highly condensed, transcriptionally iner ...
PartFourSumm_ThemesInRegulation.doc
... transription. Note that the same regulatory protein (cAMP-CAP) can make different contacts with RNA polymerase at different operons and activate transcription by different mechanisms, affecting the affinity of the polymerase for the promoter in one case and the rate of closed to open complex in the ...
... transription. Note that the same regulatory protein (cAMP-CAP) can make different contacts with RNA polymerase at different operons and activate transcription by different mechanisms, affecting the affinity of the polymerase for the promoter in one case and the rate of closed to open complex in the ...
Ch17_note_summary
... 3) Termination- stop codons (UAG, UAA, and UGA) cause that addition of water instead of amino acid, hydrolyzing and releasing the polypeptide. ...
... 3) Termination- stop codons (UAG, UAA, and UGA) cause that addition of water instead of amino acid, hydrolyzing and releasing the polypeptide. ...
Allosteric Modulation of DNA by Small Molecules
... Signals originating at the cell surface are conveyed by a complex system of interconnected signaling pathways to the nucleus. They converge at transcription factors, which in turn regulate the transcription of sets of genes that result in the gene expression. Many human diseases are caused by dysreg ...
... Signals originating at the cell surface are conveyed by a complex system of interconnected signaling pathways to the nucleus. They converge at transcription factors, which in turn regulate the transcription of sets of genes that result in the gene expression. Many human diseases are caused by dysreg ...
Protein Synthesis 1 - Transcription Translation
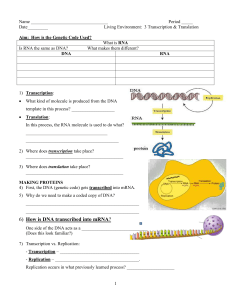
... 2) Where does transcription take place? ___________________________________________ 3) Where does translation take place? ___________________________________________ MAKING PROTEINS 4) First, the DNA (genetic code) gets transcribed into mRNA. 5) Why do we need to make a coded copy of DNA? __________ ...
... 2) Where does transcription take place? ___________________________________________ 3) Where does translation take place? ___________________________________________ MAKING PROTEINS 4) First, the DNA (genetic code) gets transcribed into mRNA. 5) Why do we need to make a coded copy of DNA? __________ ...
Lecture 14 Student Powerpoint
... transcription that involved the production of a primary transcript and the processing of this into aspects of how this is processed into an mRNA in Eukaryotes. This lecture focuses on these topics and on how signals are perceived that modulate the expression of eukaryotic genes. The next lecture wil ...
... transcription that involved the production of a primary transcript and the processing of this into aspects of how this is processed into an mRNA in Eukaryotes. This lecture focuses on these topics and on how signals are perceived that modulate the expression of eukaryotic genes. The next lecture wil ...
Gene Expression - houstonisd.org
... Regulated Gene Expression = how a cell controls the speed or amount of a gene being expressed as a protein ...
... Regulated Gene Expression = how a cell controls the speed or amount of a gene being expressed as a protein ...
Transcription Control in Eukaryotes - University of Arizona | Ecology
... Transcription control in eukaryotes is more complex than in prokaryotes, with more gene-gene interactions, presumably required to produce more different cell types in more complex organisms. We will consider some examples and models to illustrate some general principles. ...
... Transcription control in eukaryotes is more complex than in prokaryotes, with more gene-gene interactions, presumably required to produce more different cell types in more complex organisms. We will consider some examples and models to illustrate some general principles. ...
What is the difference between basal and activated transcription?
... 1. general TFs are required by all mRNA genes a. an absolute requirement b. transcription can occur alone with these factors is by definition the basal level of transcription 2. promoter-specific TFs are different for each gene 3. the promoter-specific TFs are required for maximal level of transcrip ...
... 1. general TFs are required by all mRNA genes a. an absolute requirement b. transcription can occur alone with these factors is by definition the basal level of transcription 2. promoter-specific TFs are different for each gene 3. the promoter-specific TFs are required for maximal level of transcrip ...
Our laboratory studies the regulation of gene expression in
... A second interest is the relationship between transcription and mRNA 3’-end formation. Our genetic analysis of TFIIB uncovered a novel factor, Ssu72, that we recently found to be an RNAP II CTD phosphatase with specificity for serine5-P. Interestingly, Ssu72 is an essential component of the pre-mRNA ...
... A second interest is the relationship between transcription and mRNA 3’-end formation. Our genetic analysis of TFIIB uncovered a novel factor, Ssu72, that we recently found to be an RNAP II CTD phosphatase with specificity for serine5-P. Interestingly, Ssu72 is an essential component of the pre-mRNA ...
Transcription Regulation (Prof. Fridoon)
... Many genes also have enhancer (1000 nucleotide away) where specific activators only made by certain cells can bind. ...
... Many genes also have enhancer (1000 nucleotide away) where specific activators only made by certain cells can bind. ...
Transcription factor
In molecular biology and genetics, a transcription factor (sometimes called a sequence-specific DNA-binding factor) is a protein that binds to specific DNA sequences, thereby controlling the rate of transcription of genetic information from DNA to messenger RNA. Transcription factors perform this function alone or with other proteins in a complex, by promoting (as an activator), or blocking (as a repressor) the recruitment of RNA polymerase (the enzyme that performs the transcription of genetic information from DNA to RNA) to specific genes.A defining feature of transcription factors is that they contain one or more DNA-binding domains (DBDs), which attach to specific sequences of DNA adjacent to the genes that they regulate. Additional proteins such as coactivators, chromatin remodelers, histone acetylases, deacetylases, kinases, and methylases, while also playing crucial roles in gene regulation, lack DNA-binding domains, and, therefore, are not classified as transcription factors.