分子生物學小考(一) 範圍ch3~ch7
... 6. The genetic code is said to be degenerate. This means, that (A) each codon codes for more than one amino acid (B) each anticodon can interact with many different triplet sequences in the mRNA (C) many of the amino acids are coded for by different codons (D) the code is universally used by virtual ...
... 6. The genetic code is said to be degenerate. This means, that (A) each codon codes for more than one amino acid (B) each anticodon can interact with many different triplet sequences in the mRNA (C) many of the amino acids are coded for by different codons (D) the code is universally used by virtual ...
CS 262—Lecture 1 Notes • 4-‐5 HWs, 3 late days • (Optional
... • A non-‐coding RNA called tRNA folds into itself into a complex 3D-‐structure. It has an anti-‐codon that pairs with the reverse complement codon in the mRNA and releases the correct amino acid ...
... • A non-‐coding RNA called tRNA folds into itself into a complex 3D-‐structure. It has an anti-‐codon that pairs with the reverse complement codon in the mRNA and releases the correct amino acid ...
JAK/STAT signalling • Binding of cytokines (small protein ligands) to
... Cells that communicate with each other electrically through synapses An amino acid that is often a target for kinases Ligands bind to these Created by the association of two proteins ...
... Cells that communicate with each other electrically through synapses An amino acid that is often a target for kinases Ligands bind to these Created by the association of two proteins ...
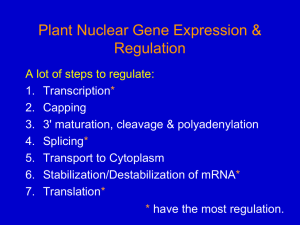
Nuclear gene expression 1
... 3. Inducible (regulated) factors: work like upstream factors but are regulatory (produced or active only at specific times/tissues); interact with enhancers or silencers. ...
... 3. Inducible (regulated) factors: work like upstream factors but are regulatory (produced or active only at specific times/tissues); interact with enhancers or silencers. ...
Pre-writing Activity for 3.05…DNA Replication For this assignment
... Pages 1 to 3 of the lesson go over the history of discovering DNA, structure of DNA, DNA replication, and DNA vs RNA. Please be sure that you take notes on these topics, as you will see this information on DBAs, module exams, and semester exams. ...
... Pages 1 to 3 of the lesson go over the history of discovering DNA, structure of DNA, DNA replication, and DNA vs RNA. Please be sure that you take notes on these topics, as you will see this information on DBAs, module exams, and semester exams. ...
Genetics
... Trait that may not be expressed Lowercase letter t= short, b=white Only expressed when there is no dominant trait present ...
... Trait that may not be expressed Lowercase letter t= short, b=white Only expressed when there is no dominant trait present ...
power point presentation
... The data collected through different approach can be used as reference to each other for possible final confidential result. ...
... The data collected through different approach can be used as reference to each other for possible final confidential result. ...
Regulation of Gene Expression in Eukaryotes
... – Function independent of orientation – Function independent of position – upstream, downstream, etc. (different than promotors‐ close to gene and only one orientation) ...
... – Function independent of orientation – Function independent of position – upstream, downstream, etc. (different than promotors‐ close to gene and only one orientation) ...
nuclear receptors - Guide to Pharmacology
... Overview:- Nuclear receptors are specialised transcription factors with commonalities of sequence and structure, which bind as homo- or heterodimers to specific consensus sequences of DNA (response elements) in the promoter region of particular target genes. They regulate (either promoting or repres ...
... Overview:- Nuclear receptors are specialised transcription factors with commonalities of sequence and structure, which bind as homo- or heterodimers to specific consensus sequences of DNA (response elements) in the promoter region of particular target genes. They regulate (either promoting or repres ...
Genetic Basis of Development
... Cells of zygote undergo rounds of mitosis to form stem cells (cells that are not yet differentiated and have the potential to develop into any type of cell) Based on their location in developing zygote, stem cells produce particular proteins (i.e. transcription factors) which tell the surrounding ce ...
... Cells of zygote undergo rounds of mitosis to form stem cells (cells that are not yet differentiated and have the potential to develop into any type of cell) Based on their location in developing zygote, stem cells produce particular proteins (i.e. transcription factors) which tell the surrounding ce ...
Transcription Worksheet and Answer Key
... The main goal of transcription is to turn ____________ into ___________. ...
... The main goal of transcription is to turn ____________ into ___________. ...
Chapter 8.4, 8.5, 8.6, 8.7 Study Guide Key terms: Ribonucleic acid
... 2. What happens to the information on a DNA molecule during transcription? 3. What are repressor proteins and where do they bind? 4. mRNA leaves the nucleus and enters the cytoplasm (with or without) a complete set of both introns and exons. (please circle the appropriate response) 5. When are intro ...
... 2. What happens to the information on a DNA molecule during transcription? 3. What are repressor proteins and where do they bind? 4. mRNA leaves the nucleus and enters the cytoplasm (with or without) a complete set of both introns and exons. (please circle the appropriate response) 5. When are intro ...
A Tale of Three Inferences
... statistical mechanical (hence concentration dependent) way. • Controversial: interaction among different transcription factor-binding events. ...
... statistical mechanical (hence concentration dependent) way. • Controversial: interaction among different transcription factor-binding events. ...
trp operon – a repressible system
... Gene regulation in eukaryotes is more complex than it is in prokaryotes because of: – the larger amount of DNA – the organization of chromatin – larger number of chromosomes – spatial separation of transcription and translation – mRNA processing – RNA stability – cellular differentiation in eukar ...
... Gene regulation in eukaryotes is more complex than it is in prokaryotes because of: – the larger amount of DNA – the organization of chromatin – larger number of chromosomes – spatial separation of transcription and translation – mRNA processing – RNA stability – cellular differentiation in eukar ...
«題目»
... Abstract The molecular basis of epigenetics involves modifications to DNA and histone proteins that associate with the regulation of gene expression but that do not result from mutation or changes to the DNA sequence. The four core histone proteins are subject to post-translational modifications, su ...
... Abstract The molecular basis of epigenetics involves modifications to DNA and histone proteins that associate with the regulation of gene expression but that do not result from mutation or changes to the DNA sequence. The four core histone proteins are subject to post-translational modifications, su ...
Signaling pathway
... w/o signal: b-catenin is continously phosphorylated, ubiquitinylated, degraded in proteasom Wnt-signal: Kinase is inhibited, non-phosphorylated b-Catenin transported into nucleus, aktivates transcription by competing of a corepressor ...
... w/o signal: b-catenin is continously phosphorylated, ubiquitinylated, degraded in proteasom Wnt-signal: Kinase is inhibited, non-phosphorylated b-Catenin transported into nucleus, aktivates transcription by competing of a corepressor ...
04/03
... regulatory proteins that bind to enhancer elements and promoterproximal elements with RNA polymerase initiates transcription at ...
... regulatory proteins that bind to enhancer elements and promoterproximal elements with RNA polymerase initiates transcription at ...
Chapter 15
... expect high or low levels of error in transcription as compared with DNA replication? Why do you think it is more important for DNA polymerase than for RNA polymerase to proofread? (Page 283) Answer: One would expect higher amounts of error in transcription over DNA replication. Proofreading is impo ...
... expect high or low levels of error in transcription as compared with DNA replication? Why do you think it is more important for DNA polymerase than for RNA polymerase to proofread? (Page 283) Answer: One would expect higher amounts of error in transcription over DNA replication. Proofreading is impo ...
Part 2
... promoter-specific transcription factors. What are some other separable domains that can be identified in these transcription factors? 14. What are the differences between Pol IIO, IIA, and IIB? Transcription and its Regulation 1. Explain the differences between activated and basal transcription both ...
... promoter-specific transcription factors. What are some other separable domains that can be identified in these transcription factors? 14. What are the differences between Pol IIO, IIA, and IIB? Transcription and its Regulation 1. Explain the differences between activated and basal transcription both ...
Control of gene expression in prokaryotes and eukaryotes
... Gene expression is transcription of DNA to make RNA and then using the RNA to make proteins. This process can’t be left on indefinitely. The turning on and off of genes is critical to the development of an organism and the organism functioning properly throughout its life. Eukaryotic control Pretran ...
... Gene expression is transcription of DNA to make RNA and then using the RNA to make proteins. This process can’t be left on indefinitely. The turning on and off of genes is critical to the development of an organism and the organism functioning properly throughout its life. Eukaryotic control Pretran ...
Synthetic approaches to transcription factor
... - Transcription factors bind to either enhancer or promoter regions of DNA adjacent to genes - Can as activators or repressors - Multiple TFs usually act on a single promoter/enhancer - Approximately 10% of genes in the human genome code for transcription factors ...
... - Transcription factors bind to either enhancer or promoter regions of DNA adjacent to genes - Can as activators or repressors - Multiple TFs usually act on a single promoter/enhancer - Approximately 10% of genes in the human genome code for transcription factors ...
Transcription Control in Eukaryotes
... Transcription control in eukaryotes is more complex than in prokaryotes, with more gene-gene interactions, presumably required to produce more different cell types in more complex organisms. We will consider some examples and models to illustrate some general principles. ...
... Transcription control in eukaryotes is more complex than in prokaryotes, with more gene-gene interactions, presumably required to produce more different cell types in more complex organisms. We will consider some examples and models to illustrate some general principles. ...
Slide 1
... cAMP receptor protein (CAP), acting as a homodimer can bind both cAMP and DNA. When glucose is absent (high cAMP state), CAP binds to its positive regulatory element increasing transcription of the lac operon 50-fold. Lac repressor is a tetrameric complex that in the absence of lactose binds tightly ...
... cAMP receptor protein (CAP), acting as a homodimer can bind both cAMP and DNA. When glucose is absent (high cAMP state), CAP binds to its positive regulatory element increasing transcription of the lac operon 50-fold. Lac repressor is a tetrameric complex that in the absence of lactose binds tightly ...
Transcription factor
In molecular biology and genetics, a transcription factor (sometimes called a sequence-specific DNA-binding factor) is a protein that binds to specific DNA sequences, thereby controlling the rate of transcription of genetic information from DNA to messenger RNA. Transcription factors perform this function alone or with other proteins in a complex, by promoting (as an activator), or blocking (as a repressor) the recruitment of RNA polymerase (the enzyme that performs the transcription of genetic information from DNA to RNA) to specific genes.A defining feature of transcription factors is that they contain one or more DNA-binding domains (DBDs), which attach to specific sequences of DNA adjacent to the genes that they regulate. Additional proteins such as coactivators, chromatin remodelers, histone acetylases, deacetylases, kinases, and methylases, while also playing crucial roles in gene regulation, lack DNA-binding domains, and, therefore, are not classified as transcription factors.