* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Nature
Signal transduction wikipedia , lookup
Drug design wikipedia , lookup
Molecular ecology wikipedia , lookup
Ligand binding assay wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Magnesium transporter wikipedia , lookup
Western blot wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Chemical synapse wikipedia , lookup
Clinical neurochemistry wikipedia , lookup
Homology modeling wikipedia , lookup
X-ray crystallography wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Metalloprotein wikipedia , lookup
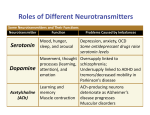
Nature 09August2007Volume448Number7154pp623726neurotransmittertransporters LeuT Nature 09 August 2007 Volume 448 Number 7154, pp623-726 Advance Online Publication LeuT 2A65 Usage of Human Famsbase for Nature 09 August 2007 Volume 448 Number 7154, pp623-726 neurotransmitter transporters on Advance Online Publication LeuT 2A65 Advance Online Publication Antidepressant binding site in a bacterial homologue of neurotransmitter transporters Satinder K. Singh, Atsuko Yamashita & Eric Gouaux Coordinates and structure factors for the alanine?sodium?clomipramine and the leucine?sodium?clomipramine, ? imipramine and ?desipramine complexes have been deposited in the Protein Data Bank under accession codes 2QEI, 2Q6H, 2Q72 and 2QB4, respectively. Status Search : Structure ID=2QEI Are you missing data updates? The PDB archive has moved to ftp://ftp.wwpdb.org. For more information click here. Status codes: PROC: to be processed; WAIT: processing started, waiting for author input to continue processing; HPUB: hold until publication; HOLD: hold until a certain date; WDRN: deposition withdrawn; AUTH: processed, waiting for author review and approval; POLC: waiting for a policy decision; REPL: author sent new coordinates, entry to be reprocessed 2QEI Crystal structure analysis of LeuT complexed with L-alanine, sodium, and clomipramine Deposition Date: 2007-06-25 Release Date: n/a Exp. Data: Structure Factors Deposited Sequence Available Status Authors NO PROC Singh, S.K., Yamashita, A., Gouaux, E. LeuT is a stable, sodium-coupled leucine transporter from the eubacterium Aquifex aeolicus and is the only member of the neurotransmitter sodium symporter (NSS or SLC6) family of secondary transporters that has so far been amenable to structural analysis17. Eukaryotic NSS counterparts include those that pump neurotransmitters such as serotonin, norepinephrine, dopamine, glycine and -aminobutyric acid (GABA) from the synapse to neuronal and glial cytoplasms, shaping the magnitude and duration of synaptic signalling18. The crystal structure of LeuT provided the first molecular glimpse into an NSS member17, but it did not yield any clues as to the atomic basis of inhibition. Motivated by this dearth of knowledge, together with the key role of NSS proteins and their antagonists in synaptic transmission, we sought to find inhibitors of LeuT and to characterize their mechanism of action by using flux, binding and crystallographic methods.We began by screening a wide variety of NSS inhibitors, including those that target the transporters of glycine (GlyT), GABA (GAT), dopamine (DAT), norepinephrine (NET), and serotonin (SERT), for their ability to inhibit the uptake of L-[3H]Leu by purified LeuT reconstituted into lipid vesicles. The tricyclic antidepressant (TCA) clomipramine was the most potent inhibitor of uptake (Fig. 1a), whereas the structurally related compounds, imipramine and desipramine, were less effective (Fig. 1a, Supplementary Fig. 1a). In dose?response experiments (Fig. 1b), clomipramine was a modestly potent inhibitor of leucine transport, with a half-maximal inhibitory concentration (IC50) approximately eightfold lower than that of imipramine (250 versus 2,090 M; Supplementary Table 1). To test whether the TCAs could displace bound leucine, we conducted radioligand binding experiments using detergent-solubilized LeuT, [3H]Leu, and potential cold competitors. Unlike amino acids such as leucine, alanine and tryptophan, none of the TCAs (at 1 mM) significantly displaced [3H]Leu from LeuT (Supplementary Fig. 1b), tentatively excluding competitive inhibition as the mechanism by which the TCAs inhibit LeuT. Structure Determination Diffraction data were collected at 110K at NSLS beamlines X26C (20-sec exposures) and X29A (5-sec exposures) or ALS beamline 8.2.2 (3-sec exposures) at an X-ray wavelength of 1.1 or 1.0 Å in a 180º or 360° sweep with 1.0º oscillation doi: 10.1038/nature06038 SUPPLEMENTARY INFORMATION www.nature.com/nature 10 and processed with HKL200031. All four inhibitor complex crystals diffracted to beyond 2.0Å resolution and were indexed in the space group C2. Unit cell dimensions varied slightly from crystal to crystal, but all were approximately a=88.0-89.8 Å, b=86.0-86.8 Å, c=80.8-81.7 Å, β=95.9-96.9º. Phases for the CMI complex were obtained via difference Fourier techniques employing the original LeuT structure (PDB ID 2A65)16 as the starting model. Subsequent model building was accomplished in O32 with the assistance of sigma-weighted 2Fo-Fc and Fo-Fc maps as well as simulated-annealing Fo-Fc omit maps. Refinement was performed with CNS33. This sequence was repeated reiteratively until the Rfactor and Rfree values converged, at which point L-leucine or L-alanine, two sodium ions, n-octylβ-D-glucopyranoside, CMI34, and water molecules were added. Refinement then progressed until Rfactor and Rfree converged again. Once the LeuT-leucine-CMI structure was completed, it was subsequently used to procure phases for the LeuTalanineCMI and LeuT-leucine-IMI35 complexes, the latter of which was then employed for the desipramine complex. For all four structures, Ramachandran geometry is excellent, with greater than 93% of the residues in the most favored regions and none in disallowed regions. For each data set, the test reflections (4.9% of the total) were selected so that they coincided with those employed in the original structure determination (PDB ID 2A65)16, which were chosen randomly. The Poisson-Boltzman equation for electrostatic calculations was solved according to the algorithm implemented in the APBS module36, 37 of PyMOL, and all structure figures were generated with PyMol38. doi: 10.1038/nature06038 SUPPLEMENTARY INFORMATION www.nature.com/nature 11 Supplementary Notes (Literature Cited) 31. Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307-326 (1997). 32. Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110-119 (1991). 33. Brunger, A. T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905-921 (1998). 34. Post, M. L. & Horn, A. S. The crystal and molecular structure of the tricyclic antidepressant chlorimipramine hydrochloride: 3-Chloro-5-(3dimethylaminopropyl)-10,11-dihydro-5H-dibenz[b,f]azepine hydrochloride. Acta Crystallogr. B 33, 2590-2595 (1977). 35. Post, M. L., Kennard, O., & Horn, A.S. The Tricyclic antidepressants: imipramine hydrochloride. The Crystal and molecular structure of 5-(3dimethylaminopropyl)-10,11-dihydro-5H-dibenz[b,f]azepine hydrochloride. Acta Crystallogr. B 31, 1008-1013 (1975). 36. Baker, N. A., Sept, D., Joseph, S., Holst, M. J. & McCammon, J. A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA 98, 10037-10041 (2001). 37. Lerner, M. G. & Carlson., H. A. (University of Michigan, Ann Arbor, 2006). 38. DeLano, W. L. (DeLano Scientific, San Carlos, CA, 2002). doi: 10.1038/nature06038 SUPPLEMENTARY INFORMATION www.nature.com/nature 12 Na(+):neurotransmitter symporter (Snf family) (2A65:A) 2A65 Title Authors Crystal structure of LEUTAA, a bacterial homolog of Na+/Cl--dependent neurotransmitter transporters Yamashita, A., Singh, S.K., Kawate, T., Jin, Y., Gouaux, E. Yamashita, A., Singh, S.K., Kawate, T., Jin, Y., Gouaux, E. Crystal Primary Citation structure of a bacterial homologue of Na(+)/Cl(-)-dependent neurotransmitter transporters. Nature v437 pp. 215-223, 2005 [ Abstract ] History Deposition Experimental Method Parameters Unit Cell Type 2005-07-01 Release 2005-08-02 X-RAY DIFFRACTION Data [ EDS ] Resolution[Å] R-Value R-Free Space Group 1.65 0.199 (work) 0.217 C 2 (C 1 2 1) Length [Å] a 87.86 b 86.31 c 81.02 Angles [°] alpha 90.00 beta 95.73 gamma 90.00 Molecular Description Polymer: 1 Asymmetric family) Molecule: Na(+):neurotransmitter symporter (Snf Chains: A Unit Classification Transport Protein Source Polymer: 1 Scientific Name: Aquifex aeolicus vf5 Expression system: Escherichia coli Ligand Chemical Component Identifier Name Formula Drug Hapten Ligand Similarity Similarity Structure Ligand Interaction BOG B-OCTYLGLUCOSIDE CL CHLORIDE ION LEU LEUCINE NA SODIUM ION GO Terms Polymer C14 H28 [ View ] [ View ] O6 [ View ] [ View ] Cl C6 H13 N [ View ] [ View ] O2 [ View ] [ View ] Na Molecular Function Biological Process Cellular Component Na(+):neurotransmitter symporter (Snf family) none (2A65:A) neurotransmitter symporter (Snf family) Na(+):neurotransmitter symporter (Snf family) (2A65:A) none none >2A65:A|PDBID|CHAIN|SEQUENCE MEVKREHWATRLGLILAMAGNAVGLGNFLRFPVQAAENGGGAFMIPYIIAFLLVGIP LMWIEWAMGRYGGAQGHGTTPAI FYLLWRNRFAKILGVFGLWIPLVVAIYYVYIESWTLGFAIKFLVGLVPEPPPNATDPDS ILRPFKEFLYSYIGVPKGDEP ILKPSLFAYIVFLITMFINVSILIRGISKGIERFAKIAMPTLFILAVFLVIRVFLLETPNGT AADGLNFLWTPDFEKLKD PGVWIAAVGQIFFTLSLGFGAIITYASYVRKDQDIVLSGLTAATLNEKAEVILGGSISIP AAVAFFGVANAVAIAKAGAF NLGFITLPAIFSQTAGGTFLGFLWFFLLFFAGLTSSIAIMQPMIAFLEDELKLSRKHAV LWTAAIVFFSAHLVMFLNKSL DEMDFWAGTIGVVFFGLTELIIFFWIFGADKAWEEINRGGIIKVPRIYYYVMRYITPA FLAVLLVVWAREYIPKIMEETH WTVWITRFYIIGLFLFLTFLVFLAERRRNHESAGTLVPR Article Title Crystal structure of a bacterial homologue of Na+/Cl--dependent neurotransmitter transporters. Abstract Na+/Cl--dependent transporters terminate synaptic transmission by using electrochemical gradients to drive the uptake of neurotransmitters, including the biogenic amines, from the synapse to the cytoplasm of neurons and glia. These transporters are the targets of therapeutic and illicit compounds, and their dysfunction has been implicated in multiple diseases of the nervous system. Here we present the crystal structure of a bacterial homologue of these transporters from Aquifex aeolicus, in complex with its substrate, leucine, and two sodium ions. The protein core consists of the first ten of twelve transmembrane segments, with segments 1-5 related to 6-10 by a pseudo-two-fold axis in the membrane plane. Leucine and the sodium ions are bound within the protein core, halfway across the membrane bilayer, in an occluded site devoid of water. The leucine and ion binding sites are defined by partially unwound transmembrane helices, with main-chain atoms and helix dipoles having key roles in substrate and ion binding. The structure reveals the architecture of this important class of transporter, illuminates the determinants of substrate binding and ion selectivity, and defines the external and internal gates. Keywords Amino Acid Sequence, Bacteria, Bacterial Proteins, Binding Sites, Biological Transport, Chlorides, Crystallography, X-Ray, Hydrophobicity, Leucine, Membrane Transport Proteins, Models, Molecular, Molecular Sequence Data, Neurotransmitter Agents, Sequence Alignment, Sodium, Structure-Activity Relationship, Water Na(+):neurotransmitter symporter (Snf family) Aquifex aeolicus Authors Yamashita, A., Singh, S.K., Kawate, T., Jin, Y., Gouaux, E. Organizational Affiliation Department of Biochemistry and Molecular Biophysics and. Journal Nature v437 pp. 215-23, 2005 Pubmed ID