* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Example 2. - the Gene Ontology Consortium Wiki
Epigenetics in stem-cell differentiation wikipedia , lookup
Epitranscriptome wikipedia , lookup
DNA vaccination wikipedia , lookup
Gene expression programming wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Primary transcript wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Gene expression profiling wikipedia , lookup
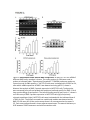
General information that should be discussed and agreed for guidelines For a long time scientists have agreed that the level of an mRNA does not directly correlate with the level of a protein and this is, for the most part, reflected in the way that curators annotate experimental data. Similarly, the level of mRNA expression (which can be modulated e.g. by microRNAs) is not necessarily equivalent to the level of transcription; however, curators sometimes do not capture this distinction, especially when they take into consideration authors’ interpretation of results, i.e. the authors’ intent. With this in mind we would like to have an open discussion about how mRNA expression data is annotated. Objectives: (1) To understand how curators annotate non-high-throughput mRNA expression data; (2) To discuss how to improve consistency when annotating mRNA expression experiments. Example 1. Re: PMID: 19805038, Figure 2. Curator note: Human T-box protein 21 (T-bet) summary: Transcription factor that controls the expression of the TH1 cytokine, interferon-gamma. Initiates TH1 lineage development from naive TH precursor cells both by activating TH1 genetic programs and by repressing the opposing TH2 programs. (http://www.uniprot.org/uniprot/Q9UL17). In this example we are interested in how you would annotate the regulation of expression of targets of T-box protein 21 (T-bet). Abstract Upon detection of antigen, CD4+ T helper (Th) cells can differentiate into a number of effector types that tailor the immune response to different pathogens. Alternative Th1 and Th2 cell fates are specified by the transcription factors T-bet and GATA-3, respectively. Only a handful of target genes are known for these two factors and because of this, the mechanism through which T-bet and GATA-3 induce differentiation toward alternative cell fates is not fully understood. Here, we provide a genomic map of T-bet and GATA-3 binding in primary human T cells and identify their target genes, most of which are previously unknown. In Th1 cells, T-bet associates with genes of diverse function, including those with roles in transcriptional regulation, chemotaxis and adhesion. GATA-3 occupies genes in both Th1 and Th2 cells and, unexpectedly, shares a large proportion of targets with T-bet. Re-complementation of T-bet alters the expression of these genes in a manner that mirrors their differential expression between Th1 and Th2 lineages. These data show that the choice between Th1 and Th2 lineage commitment is the result of the opposing action of T-bet and GATA-3 at a shared set of target genes and may provide a general paradigm for the interaction of lineagespecifying transcription factors. Figure 2. T-bet activates the expression of its target genes. (A) Relative levels of expression of T-bet target genes in primary stimulated Th1 cells compared with Th2 cells (column 1) and in secondary stimulated Th1 compared with Th2 cells (column 2). Each row represents a single gene. Genes are ordered by their relative expression level in Th1 cells versus Th2 cells and this average expression ratio is shown on the left hand side. The colors represent relative gene expression levels between Th1 and Th2 cells, with shades of red indicating higher expression in Th1 cells, black equal expression levels and shades of green indicating higher expression in Th2 cells. (B and E) T-bet ChIP signals at NKG7 and CCL4, in human Th1 cells (green) and Th2 cells (blue), and signal from an IgG control IP in Th1 cells (black). Details as for Fig. 1B. (C and F) Real time PCR for NKG7 and CCL4 in WT (open bars) and T-bet−/− (filled bars) murine T cells. Cells were unstimulated (u/s), stimulated with anti-CD3 antibodies (αCD3), or stimulated with anti-CD3 antibodies and anti-CD28 antibodies (αCD3/28). RNA abundance is relative β-actin RNA. (D and G) Real time PCR for NKG7 and CCL4 in CD4+ T cells from T-bet−/−× IFN-γ−/− murine CD4+ T cells transduced with empty vector (open bars) or T-bet (filled bars) expressing retrovirus. (H) Percentage of human Th1 genes whose orthologs are upregulated (at least 1.5-fold, black bars) or downregulated (white bars) in murine Tbet−/−× IFN-γ−/− CD4+ cells upon expression of exogenous T-bet. Th1 genes were defined as those expressed at least 2-fold or higher in Th1 cells than Th2 cells and divided into those bound by T-bet or those not bound by T-bet. Example 2. Re: PMID: 24296617, Figure 2. Curator note: miR-19b is predicted to target CTGF for silencing (Fig. 3a not shown here). Human AngII summary: A peptide hormone that is an essential component of the renin-angiotensin system, a potent regulator of blood pressure, body fluid and electrolyte homeostasis. AngII acts directly on vascular smooth muscle as a potent vasoconstrictor (Full UniProt description: http://www.uniprot.org/uniprot/P01019). In this example we are interested in how you would annotate the regulation of CTGF expression by AngII. Abstract Objectives: The present study was designed to decipher the molecular mechanisms underlying angiotensin (Ang) II-induced overexpression of connective tissue growth factor (CTGF) in cultured cardiomyocytes. Methods: Cardiomyocytes isolated from 1- to 3-day-old neonatal rats were cultured and treated with 100 nM [human] Ang II with or without pretreatment with 10 nM telmisartan, an Ang II type 1 receptor antagonist. The role of microRNA (miR)-19b in the regulation of Ang II-induced CTGF expression was evaluated in cultured cardiomyocytes with quantitative real-time reverse transcription polymerase chain reaction and Western blot analysis. Results: We provide several lines of evidence to show that miR-19b contributes to the Ang II-induced overexpression of CTGF in cultured cardiomyocytes. Firstly, administration of Ang II decreased the level of miR-19b dramatically (p < 0.05 vs. control), which was abolished by telmisartan. Secondly, Ang II increased the level of CTGF significantly (p < 0.05 vs. control), which was also prevented by pretreatment with telmisartan. Thirdly, overexpression of miR-19b decreased CTGF levels (p < 0.05 vs. control). Finally, transfection of miR-19b into cardiomyocytes prevented the upregulation of CTGF induced by Ang II. Conclusion: Downregulation of miR-19b contributes to Ang II-induced overexpression of CTGF in cultured cardiomyocytes. “As shown in figure 2 a and b, CTGF mRNA expression was significantly increased after Ang II stimulation (p < 0.05 vs. control group). When compared with the Ang II-treated group, the induction of CTGF gene expression was prevented by preincubation with telmisartan (p < 0.05). The result of Western blot showed that Ang [angiotensin] II regulated the expression of CTGF [connective tissue growth factor] at protein level, and preincubation with telmisartan prevented the effect of Ang II on CTGF expression (fig. 2 c, d). These results suggest a mechanism that mediates the regulation of expression of CTGF genes by Ang II via an Ang II type 1 receptor-dependent (AT 1 R-dependent) pathway.” Example 3. Re: PMID:20675382, Figure 2. Curator note: CYR61 is known to bind the extracellular matrix, heparin and integrins. CYR61-mediated gene regulation is dependent on heparin-binding. (Full UniProt description: http://www.uniprot.org/uniprot/O00622). In this example we are interested in how you would annotate the regulation of BMP-2 expression by CYR61 "CYR61 Induced BMP-2 Production in Osteoblast Cells. Given the crucial role of BMPs in osteoblastic differentiation, we tested whether CYR61 mediated alteration of osteoblast proliferation and differentiation through regulation of BMP expression. According to Cheng et al. (43), BMP-2, -6, and -9 may be the most potent molecules that induce osteoblast lineage-specific differentiation. Therefore, we explored possible target gene expression during rCYR61-induced osteoblastic differentiation. We examined expression levels of members of the BMP family by RTPCR in MC3T3-E1 osteoblast cells in response to recombinant CYR61 (rCYR61) treatment. Treatment with rCYR61 stimulated BMP-2 expression in a time-dependent manner by RT-PCR and Western blot analysis (Fig. 2, A and D). A significant increase in BMP-2 mRNA could be detected as early as 2 h after rCYR61 treatment. Elevation of BMP-2 mRNA was maximal at 8 h and lasted until 24 h after treatment. Expression level of other BMP mRNAs, such as BMP-4, -6, -7, and -9, were not significantly affected by rCYR61 (Fig. 2A). In addition, the stimulatory effect of rCYR61 on BMP-2 expression was dose-dependent in the range between 10 and 200 ng/ml (Fig. 2, B and C). The BMP-2 protein expression level in response to rCYR61 treatment was increased at 4~24 h after treatment (Fig. 2D). Furthermore, we found rCYR61 induced BMP-2 secretion into cultured medium of both MC3T3-E1 cells and primary osteoblasts by BMP-2 ELISA assay (Fig. 2E). These data indicate that rCYR61 induces expression of BMP-2 in osteoblast cells in a time- and dose-dependent manner."... "we hypothesized that, as a positive extracellular signaling protein, CYR61 controls osteoblast functions by regulating BMP2 gene expression in osteoblasts. This study was designed to test this hypothesis and also determine the precise signaling mechanisms involved in CYR61 regulation of BMP2 transcription in osteoblasts.” Figure 2. Recombinant CYR61 induces BMP-2 expression. A and B, RT-PCR for mRNA of different BMP family members is shown. The coding regions of cDNA were used as probes as indicated under “Experimental Procedures.” A GAPDH probe was used as an internal control for RNA quantity. C, MC3T3-E1 cells were incubated with rCYR61 for 8 h, after which mRNA expression of BMP-2 was determined using qPCR. D, shown is Western blot analysis of BMP-2 protein expression in MC3T3-E1 cells. Total proteins were extracted from cells and probed with polyclonal antibody specific for BMP-2. Each lane contains 80 μg of total protein. The internal loading control was β-actin. E, shown is an ELISA assay of BMP-2 protein expression in MC3T3-E1 cells (left) and primary cultured osteoblasts (right). MC3T3-E1 cells were incubated with the indicated dose of rCYR61 for 24 h. The protein secreted into conditioned medium was determined by BMP-2 ELISA assay kit. All the results except those in D are expressed as the means ± S.E. Each assay was performed in three separate experiments. The asterisk indicates p < 0.05 between rCYR61 treatment and vehicle treatment cells.