* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download US Tomato sequencing project http://sgn.cornell.edu/
Survey
Document related concepts
Genome (book) wikipedia , lookup
Microevolution wikipedia , lookup
Designer baby wikipedia , lookup
Neocentromere wikipedia , lookup
Human genome wikipedia , lookup
Genome evolution wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Pathogenomics wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Whole genome sequencing wikipedia , lookup
Human Genome Project wikipedia , lookup
Star Wars: Episode II – Attack of the Clones wikipedia , lookup
Transcript
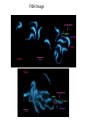
US Tomato sequencing project update http://sgn.cornell.edu/ January 14, 2007 US Tomato Genome sequencing ● BAC libraries ● ● Made two BAC libraries (EcoRI & MboI) in addition to HindIII library BAC end sequence 400,000 BAC end sequence reads 340,000 high quality insert sequences Chromosomes to be sequenced 1, 10, 11 Sequenced 17 full BACs to date > 40 successful FISH hybridizations $1.8 million in support from NSF (Fall, 06) Pending proposal for full sequencing of Chromosomes 1, 10, 11 BAC libraries and BAC end sequences Library Name / enzyme HindIII MboI EcoRI Sheared library Total Number of Approx number of Cloning Vector clones clones seqenced 129024 76000 pBeloBAC11 100,000 50688 25344 pEC BAC I 50,000 50,000 75000 25344 pIndigoBAC-5 N.A. 4800 PUC18-SW Additional ordered libraries: S. cheesmannii S. pennellii HindIII HindIII S. lycopersicum Sau3A S. lycopersicum Sau3A pBeloBAC11 pBeloBAC11 100,000 clones 100,000 clones cosmid cosmid 200,000 clones 20 kb avg. >100,000 clones > 20 kb avg. S. lycopersicum sheared fosmid >100kb avg. >100kb avg. >150,000 clones 40 kb avg. (400,000 target) SSR125 cLEC7H4 SSR331 SSR580 cLET1I9 SSR103 T1201 T634 cLER17N11 TG154 Fw2.2 T1480 T1566 T347 CT9 T1665 CT38 SSR50 T147 SSR32 T1494 SSR26 T562 SSR5 SSR349A T697 SSR605 T1706 T1117 TG31 SSR57 SSR356 CT255 SSR96 SSR66 SSR40 ● SSR586 ● cLEC7P21 T1616 Overgo Project anchor tomato BACs/contigs on the highly saturated genetic map (F2.2000) identify the minimum tiling path of BAC clones for BAC-by-BAC sequencing FISH Image Bioinformatics ● BAC registry database ● SGN Data repository ● Central database at SGN that keeps track of the status of every BAC sequenced in the project All sequences, including all primary data (chromatograms and assemblies) are uploaded to the central data repository Participation in ITAG annotation Structural Annotation pipeline Functional Annotation pipeline Hetero/euchromatin BAC repeat annotation Euchromatin: Gene rich, repeat poor Genes Heterochromatin: Gene poor, repeat rich (red) Repeats Future plans ● ● ● ● Complete and End-sequence Fosmid library (400,000 clones) Full sequences of chromosome 1, 10 & 11 (estimated 550 BACs) Support international project partners with BAC libraries and FISH (10 hybes/country) Continue to run a central bioinformatics hub for data deposition (SGN), project tracking and running shared annotation pipeline Acknowledgments Steven Tanksley Yimin Xu Nancy Eanetta Jim Giovannoni Ruth White Julia Vrebalov Joyce van Eck Stephen Stack Suzanne Royer SGN: Lukas Mueller Naama Menda Rob Buels Marty Kreuter Chenwei Lin John Binns Beth Skwarecki