* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Network (Reticulate) Evolution: Biology, Models, and
Maximum parsimony (phylogenetics) wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Gene expression profiling wikipedia , lookup
Pathogenomics wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Gene therapy wikipedia , lookup
Gene desert wikipedia , lookup
Gene nomenclature wikipedia , lookup
DNA barcoding wikipedia , lookup
Population genetics wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Genetic engineering wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
History of genetic engineering wikipedia , lookup
Genome (book) wikipedia , lookup
Helitron (biology) wikipedia , lookup
Hybrid (biology) wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Genome evolution wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Designer baby wikipedia , lookup
Gene expression programming wikipedia , lookup
Computational phylogenetics wikipedia , lookup
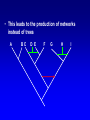
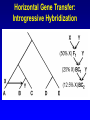
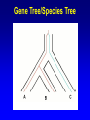
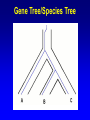
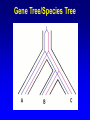
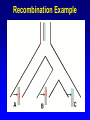
Network (Reticulate) Evolution: Biology, Models, and Algorithms C. Randal Linder*, Bernard M.E. Moret† *University of Texas at Austin (currently the Program for Evolutionary Dynamics, Harvard University) †University of New Mexico Purpose of Tutorial • Familiarize you with the nature of reticulation in biology, especially hybrid speciation • Discuss the implications of reticulation for our understanding of evolution • Present currently available methods for simulating, detecting and reconstructing reticulation • Consider deficiencies of the current methods Overview of Reticulation in Biology • What happens at the genetic level? • How does it relate to population genetic processes? – In particular, what processes can give the appearance of species level reticulation • How can we detect it? • How can we reconstruct it? • What biological tools need to be in place to generate the requisite data? Idealized Nature • Wouldn’t it be nice if… – Sexual creatures would just behave themselves – Asexual lineages would keep their pseudopods to themselves Then we could stick with bifurcating graphs (trees) to properly describe the evolutionary history of organismal lineages A B C D E F G H Unruly Nature Whatever is not forbidden will occur. -- Gerald Myers (ca 1980) In Other Words • Nature does not care about our nice systems • Rather, the only rule is: – If a set of genes can be brought together in a cell, survival and reproduction will be determined by the phenotype produced in the environment of the organism. • If the organism can survive and reproduce as well as or better than its competitors, it “works” no matter the mating/process that produced it Therefore • Some “species” are able to interbreed or exchange genes in ways that violate “normal” notions about species and speciation • Reticulation is violation of the independence of each evolutionary lineage – Instead of bifurcation, lineages can mix and produce new lineages • This leads to the production of networks instead of trees A BC D E F G H I Molecular phylogeneticists will have failed to find the “true tree,” not because their methods are inadequate or because they have chosen the wrong genes, but because the history of life cannot properly be represented as a tree. --Ford Doolittle Before Reticulation • Paradoxically, I’ll begin with non-reticulate evolution • Bifurcating evolution (and sometimes hard polytomies) – Evolutionary lineages split and evolve independently from one another Before Reticulation • Key Evolutionary Insight: Because all evolution is a product of change from one generation to the next, the information must initially change in some form of bifurcating process. agct acct agct gcct acct gcct gact agct agat With Reticulation • The end result is admixture of different evolutionary histories agct acat agct acat acat acct agct agct Levels of Reticulation • Life is organized hierarchically and so reticulation can occur at different levels – Chromosomal (meiotic recombination) – Population (sexual recombination) – Species (interspecific hybridization and horizontal gene transfer) Levels of Reticulation • Chromosomal (meiotic recombination) Levels of Reticulation Population (sexual recombination of haploid genomes) Levels of Reticulation • Species (hybridization and gene transfer) Levels Nested within Levels Areas of Biological Research • Most of the work on reticulation has been done at the population genetic level – A great deal of work on recombination, especially meiotic recombination • Hybrid speciation and lateral gene transfer are less well studied – Intersect with the population genetic perspective – Will talk about this a bit later and from other speakers Types of Hybrid Speciation • Allopolyploidization: each parent of the hybrid contributes it’s entire nuclear genome (usually uniparental inheritance of the organelles) – Parents needn’t have the same number of chromosomes Types of Hybrid Speciation • Diploid (Homoploid) Hybridization: each parent contributes half of its diploid chromosome set, as it would with normal sex. – Parents almost always have the same number of chromosomes Types of Hybrid Speciation • Autopolyploidization: a doubling of the diploid chromosome number in a single species – From a biological and topological perspective, could be considered a type of bifurcating speciation Horizontal Gene Transfer • Hybridization between lineages, but an independent lineage is not produced – Hybrids backcross to one or both parents allowing introgression of genes between “species” • Genes are moved between lineages by a third party (vector), e.g., a virus Horizontal Gene Transfer: Introgressive Hybridization Horizontal Gene Transfer: Genome Capture • A complete organellar genome is transferred by hybridization Horizontal Gene Transfer: Bacterial Sex • Genetic material is moved by conjugation between compatible bacteria Bacteria: Promiscuous DNA Sharers • Lawrence, Ochman estimated that 755 of 4,288 ORFs in E. coli were from at least 234 lateral gene transfer events (Proc. Natl Acad. Sci. USA 95, 9413-9417 (1998) ) • General evidence: Horizontal Gene Transfer: Exchange by a Vector • Genetic material is moved by a third party such as a virus or a combination of organisms, e.g., mosquito and protozoan. Neworks Have Incongruent Trees Within Them Reticulation Events Have Incongruent Trees Within Them Reticulation Events Have Incongruent Trees Within Them Fundamental Insight • At the lowest possible level (individual DNA nucleotides on a single DNA strand) all evolution is ultimately tree-like. How Might We Detect Reticulation? • Fundamentally, reticulation is a mixing of different evolutionary signals. Therefore: – The signal from a genome that has experienced reticulation will be an “average” of its parents (Median approach) – Unrecombined stretches of DNA will have a signal that comes from one parent. (Incongruence approach) • Will see both approaches in methods for detection and reconstruction Evolutionary Events that Mimic Species-Level Reticulation • Lineage Sorting (gene tree/species tree problem) • Reticulation at lower levels, e.g., meiotic recombination Evolutionary Events that Mimic Species-Level Reticulation • Lineage Sorting (gene tree/species tree problem) – When reconstructing a species-level phylogeny using DNA sequence information we are actually reconstructing a gene tree. – Ancient alleles (alleles arising prior to some monophyletic group) may not be inherited by all species. – In essence, it is either a sampling problem or an irretrievable information loss problem. Gene Tree/Species Tree Gene Tree/Species Tree • All of the versions of a gene from a single common history (everything that is the same color) are referred to as orthologues. • Versions of a gene from a duplication event or the production of a new allele are paralogues Gene Tree/Species Tree Gene Tree/Species Tree Gene Tree/Species Tree Gene Tree/Species Tree Gene Tree/Species Tree Gene Tree/Species Tree Gene Tree/Species Tree • Under a molecular clock, it is possible to detect the difference between incongruence due to hybridization and to a gene tree/species tree sampling problem. •GT/ST incongruences will occur at different depths. Evolutionary Events that Mimic Species-Level Reticulation • Reticulation at lower levels, e.g., meiotic recombination – Recombination can lead to loss of an allele for a lineage in a particular region of DNA essentially giving rise to a lineage sorting problem. Recombination Example Second Key Insight • Events that masquerade as species-level reticulate evolution are always the product of either true data loss or inadequate sampling. – Here, we encounter the importance of a population genetic perspective in phylogenetics. Given the problem of misleading signals, how can we distinguish true specieslevel reticulation from reticulation at other levels, simple data loss, and inadequate sampling? Possible Solution • Increase the number of individuals sampled from a species/population and the number of markers. • Therefore, must take a multiple marker approach to recovering the specieslevel relationships – Data loss and lower level reticulation events should almost always act randomly with respect to which phylogeny is favored – Species-level reticulation will be biased toward a particular interpretation Practical Concerns • Practical problems (for biologists): – Cost – Time – Lack of prior knowledge that all of the orthologues are there to be found Caveats • Reticulation events that quickly follow speciation may not be detectable • Ancient reticulation events may not be recoverable • The computational requirements to detect and reconstruct reticulation may be considerable • We may have to rethink our ideas of species (levels/units of speciation) Assembling the Network of Life: ANOL