* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Course Outline - North Carolina State University
Epigenetics of human development wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Hardy–Weinberg principle wikipedia , lookup
Gene expression profiling wikipedia , lookup
Pharmacogenomics wikipedia , lookup
Polymorphism (biology) wikipedia , lookup
Public health genomics wikipedia , lookup
Minimal genome wikipedia , lookup
History of genetic engineering wikipedia , lookup
Genomic imprinting wikipedia , lookup
Genetic drift wikipedia , lookup
Genetics and archaeogenetics of South Asia wikipedia , lookup
Genome evolution wikipedia , lookup
Genome (book) wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Human genetic variation wikipedia , lookup
Medical genetics wikipedia , lookup
Dominance (genetics) wikipedia , lookup
Population genetics wikipedia , lookup
Designer baby wikipedia , lookup
Microevolution wikipedia , lookup
Behavioural genetics wikipedia , lookup
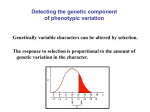
Introductory Genetics 1B Heritability and Complex Traits Greg Gibson Department of Genetics North Carolina State University Course Outline 1A. 1B. 2A. 2B. 3A. 3B. 4A. 4B. 5A. 5B. Mendelian Genetics (JA) Heritability and Complex Traits (GG) The Central Dogma and Gene Structure (JA) Gene Regulation (GG) Linkage Mapping (GG) QTL Mapping (JA) Genome Sequencing (JA) Genome Annotation (GG) Microarrays (GG) Association Studies (JA) Introduction • Genetic analysis of common human diseases, animal and plant breeding, and evolutionary biology all require the tools of quantitative genetics. • Quantitative genetics is the study of the variation in continuous traits. • Small effects of many individual genes add together and interact with the environment, to produce natural variation • Modern molecular quantitative genetics is focused on identifying the underlying genes and describing how variation at the DNA level translates into phenotypic variation Oligogenic effects 1 Locus Frequency Frequency 2 Loci 1 2 - + - 1 + + 1 4 - - - + - - Frequency 3 Loci 1 - - - - - 6 15 20 15 + + - - - - - + + + - - + - - + - - + - + + - + - - + + + + - - 6 1 + + + + + - - + + + + + + + - + + + - 6 + + + + - 4 + + + - 1 + + + + Types of Complex Trait • Continuous traits (eg height, IQ) • Meristic traits (eg number of petals, whiskers) • Discrete traits (eg disease status, color) • Threshold-dependent traits Heritability • Heritability is the proportion of phenotypic variation in a population that is due to genotypic variation. • Heritability describes the relative contributions of genotype and environment to phenotypic variation. • Heritability is a measure of a population, not an individual, and does not say anything about the genetic difference between populations. • There are three common approaches to measuring heritability. Parent-Offspring Regression Offspring Phenotype 6 5 4 3 2 1 1 2 3 4 Parental Phenotype 5 6 Variance among Lines High heritability 6 6 5 5 4 4 Phenotype Phenotype Low heritability 3 3 2 2 1 1 1 2 3 Line Number 4 1 2 3 Line Number 4 Realized heritability Frequency Initial population Selected sample O R S R Progeny of selected sample Phenotype S Polygenes • The number of genes that affect a trait can be estimated from the variance segregating in an F2 generation. • There is a wide range in the number of genes that affect traits. • As the number of genes influencing a trait increases, the relative number of individuals with extreme phenotypes decreases. • Transgressive segregation occurs when grandchildren have more extreme phenotypes than either grandparent. Simple additive model ¼ AA (0) ½ Aa (1) ¼ aa (2) ¼ BB (0) 1/16 (0) 1/8 (1) 1/16 (2) ½ Bb (1) 1/8 (1) 1/4 (2) 1/8 (3) ¼ Bb (2) 1/16 (2) 1/8 (3) 1/16 (4) Polygenic effects Frequency Parental samples Frequency F2 samples Frequency F1 sample Phenotype Transgressive segregation Parental samples Frequency A. - ; + + ; -- ; - + + ; - ; ++ ; + + B. F2 F1 F2 samples Frequency P2 Frequency P1 25 35 45 55 65 75 85 95 105 115 Plant height (cm) - ; - ; -- ; - + + Phenotype ; + + ; ++ ; + + VP = VA + VD + VI + VGxE + VE • Loci are said to have Additive effects if the contributions of each individual allele can simply be added algebraically to arrive at a prediction of a phenotype given a genotype. • Dominance refers to the observation that heterozygotes resemble one class of homozygotes more than the other. • Epistasis refers to a locus-by-locus Interaction, such as when alleles at two loci antagonize or synergize with one another. • VE is the environmental variance Growth Rate (mg/h) Norm of Reaction 2 1 0 11 17 23 29 Growth Temperature (oC) 35 43 Mean phenotype Dominance ratio 20 18 16 14 12 10 8 6 4 2 0 bb Bb a d d = 16-10 = 6 a = 18-10 = 8 0 Expected mid-value = (18+2)/2 = 10 BB -a 0 1 Number of “b” alleles 2 Epistasis OBSERVED 38 37 36 35 34 33 32 31 EXPECTED 38 37 36 35 34 33 32 31 BB Bb bb AA Aa BB Bb bb aa AA EPISTATIC DEVIATIONS 1.00 0.75 0.50 0.25 0.00 - 0.25 - 0.50 - 0.75 - 1.00 Bb bb BB AA Aa aa Aa aa