* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Document
Quantitative trait locus wikipedia , lookup
History of genetic engineering wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Designer baby wikipedia , lookup
Oncogenomics wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Frameshift mutation wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Genome editing wikipedia , lookup
Dominance (genetics) wikipedia , lookup
Cell-free fetal DNA wikipedia , lookup
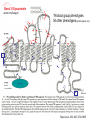
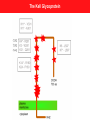
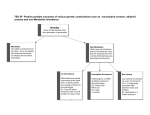
Abstract HGVS meeting Toronto, October 26, 2004 Phenotypes vs Genotypes in the World of Blood Group Antigens Olga O. Blumenfeld1 and Santosh K. Patnaik2 Deparments of Biochemi stry1 and Cell Biology2. Albert Einstein Coll ege of Medicine. New York, NY 10461 Blood group antigens are proteins, gly cans or glycoli pids, of a variety of functions, whose comm on feature is that all are expressed on the surface of red cell s and are polym orphic in the population. The hallm ark of each antigen is an epitope or a li near or spatially arranged sequence of ami no acids, or a carbohydrate sequence which, due to its variant nature, can be recognized as non-self by the imm une system. The science (art) of serology is based on this recognition, and its goal is to decipher and assign blood group phenotypes using antibodies to the polym orphic epitopes as too ls. A blood group system is a set of variant antigens encoded by all eles of a single locus, each expressing a related form of a comm on blood group phenotype. The Blood Group Antigen Gene Mutation Database* documents 39 gene loci encoding 29 blood group systems and comprising a tot al of 688 all eles that result in surface expression of at least 400 diff erent blood group phenotypes. Here we exami ne the correlation between the genotype, the structure of the all ele and the blood group phenotype. This is a rare example in which a direct correlation between a DNA alteration and a single physiologic function (antibody response) can be establi shed, with modifi er genes or environmental factors playing a mi nim al role. In the database, DNA alterations were documented in donors who were selected for study on the basis of a variant blood group phenotype. Thus an alteration of the epitopic and/or coding regions was expected. As generally observed for many documented human sequence variations, among tot al DNA alterations, single nucleotide mutations predomi nated (sense ~8%; missense ~ 50%, nonsense ~6%). Nonsense mutations and small deletions, insertions, or spli ce site alterations, often accompanied by additional upstream or downstream mutations, gave rise to a number of related all eles whose products were trunca ted or defective and resulted, directly or indirectly (inactive glycosyltransferases), in the absence of epitopes from the cell surface. Thus, for several blood group systems such diff erent all eles resulted in the null phenotype (for example, the O phenotype of the ABO system). Gene rearrangements based on different mechanisms (gene conversions, unequal recombinations) could also give rise to the same epitopic sequence and the same phenotype that specifi ed diff erent hybrid all eles (the MN system). In contrast, in most instances, mi ssense mutations li nked to the epitope, gave rise to variant phenotypes each characteristic of a specifi c mutation and the ami no acid change (Rh, Diego, Kell , others). This was observed whether the protein exhibited a single or multiple epitopes and the effect of the mutation could be direct or indirect (affecting level of cell surface expression). The majority of DNA alterations had no apparent effect on the function of the erythrocyte. The subtle relationships among a single ami no acid replacement affecting the epitopic region, the immune response and the abil ity to detect it, become apparent from a survey of different blood group systems documented in the database. *http://www. bioc.aecom.yu.edu/bgmut/index.htm Phenotypes in the World of Blood Group Antigens documented in the Database of DNA Variation in Genes Encoding Blood Group Antigens Olga O. Blumenfeld & Santosh K. Patnaik Departments of Biochemistry (OOB) & Cell Biology (SKP) Albert Einstein College of Medicine, New York www.bioc.aecom.yu.edu/bgmut/index.htm Blood Group Antigens proteins, glycans or glycolipids variety of functions expressed at the surface of red cells polymorphic in the population Blood Group System A set of variant antigens resulting from alleles of a single locus, each defining a common serological phenotype. Summary: 29 blood group systems, 40 genes, 707 alleles Also detailed: non-human counterparts for H/h, MN, Rh System Locus ABO Chido- Rodgers ABO enzyme 115 C4A, factor 7+ C4B AQP1 channel 7 DAF receptor 13 SLC4A1 exchanger 78 DO unknown 9 FY receptor 7 GYPC structure 9 AQP3 channel 2 FUT1, enzymes 57 FUT2 GCN2 enzyme 8 (IGnT) CD44 adhesion 2 SEMA7A signaling 0 KEL, enzyme 67 XK SLC14A1 transport 8 CR1 receptor 24+ Colton Cromer Diego Dombrock Duffy Gerbich (Ge) GIL H/h I Indian (IN) JMH Kell (with Kx) Kidd Knops Funcion Alleles System LandsteinerWeiner Lewis Lutheran MNS OK P-related RAPH-MER2 Rh Scianna Xg YT Locus Function Alleles ICAM4 adhesion (LW) FUT3, enzymes FUT6, FUT7 LU adhesion GYPA, unknown GYPB, GYPE BSG adhesion A4GALT, enzymes B3GALT3 CD151 RHCE, transport RHD, RHCG RHAG, RHBG ERMAP adhesion XG, adhesion CD99 (MIC2) ACHE enzyme 3 36 16 43 5 27 3 126 4 0 4 Summary of DNA alterations Locus Sens e Mis sense Nonsense Spli cing Inse rtion Deletion Rerrange ments Gross Recur rent identical alt erations at same sit es ABO A4GALT CR1 KEL XK FUT1 FUT2 FUT3 17 3 1 2 1 3 - 29 4 16 26 2 16 6 11 2 1 6 6 4 3 - 3 5 - 1 3 1 1 1 - 2 4 6 3 2 - 22 - 8 - FUT6 GYPA GYPB RHCE RHD RHAG SLC4A1 BGMUT db total HGMD total 7 2 2 2 - 9 14 7 13 45 5 48 1 1 3 4 1 1 6 3 19 1 1 3 7 19 2 2 12 33 243 243 183 24 92 2 2 12 3207 2093 5498 340 1827 312 19368 1. For comparison, nu mber of corresponding en tries in H GMD is shown in pa renthes es; n/a: no t ava il able in HG MD. Not includ ed: regu latory mutations , small inde ls , repeat variations , gross insertions & duplic ations. Total docu mented for 1338 gen es 2. Minimal number; approximate nu mbers as breakpoints for the p roposed intragen ic rearrange ments and g ene conv ersions are unknown 3. Rough e stimate of all numb ers because o f a variety o f nu merous gene rearrang ements Total DNA chang es1 19 4 5 6 Recur rent dif ferent alt erations at same sit es 3 2 - 13 - - 18 (5) 41 (1) 35 (n/a) 37 (12) 77 (8) 14 (11) 74 (78) 497 53 (20) 15 (n/a) 17 (n/a) 38 (14) 28 (n/a) 25 (25) 14 (15) 11 (10) 32333 Phenotype vs genotype A number of alleles give rise to the same blood group phenotype Silencing mutations nonsense, deletions, insertions, splicing, regulatory regions rearrangements, gross deletions (null phenotype in nearly all systems) Kell, Rh, Diego, ABO, others Gene rearrangements GYPA, Rh ex. Sta - 7 genotypes; RH neg - 17 genotypes A single allele gives rise to a unique blood group phenotype Missense mutations Kell, Diego, RH, Duffy, ABO and others Gene rearrangements GYPA, Miltenberger series; Rh, weak D(Du), DAU & others Same Blood Group Phenotype, Multiple Genotypes Kell null 1 phenotype 11 genotypes 7 nonsense 3 splicing 1 deletion Same Blood Group Phenotype, Multiple Genotypes O gene-null Same deletion in many alleles (261delG in 38 of 43 O null alleles) Band 3 Glycoprotein anion exchanger 19 blood group phenotypes 54 other phenotypes (spherocytosis, etc.) del 19 variants Binds to rbc cytoskeleton del anion ex. Popov et al. JBC.1997,272,18325 Multiple Blood Group Phenotypes, Multiple Genotypes Diego 19 of 19 alleles Sites and distribution of alterations vs location of epitopes: Kell (KEL) Each polymorphic site can be assigned to a different Kell antigen 24 missense mutations at positions in extracellular domains 7 nonsense; 3 splicing ;1 del Sets of polymorphic residues Each expressed in different individuals at different frequencies. Common phenotype K-1, 2*; -3, -21, 4*; -6, 7*; 11*, -17; 14*, -24; 10, 5, 12, 13, 16, 18, 20, 22, -23 Known antithetical partners K2/K1; K4/K3/K21; K7/K6; K11/K17 The Kell Glycoprotein Single Blood Group Phenotype, Single Genotype Kell 22 of 24 alleles note note note Same Blood Group Phenotype, Multiple Genotypes: Examples in the KEL blood group system Single Blood Group Phenotype, Single Genotype Rh Examples of genotypes vs phenotypes due to DNA rearrangements in GYPA family Gene Sequence Phenotype GYPA GYPB GYPB ex3 EET ex4 GERVQL ex2 QTN ex4 GETGQL ex2 QTN ex4 GEMGQL wild type wild type s wild type S hyb.A-Bs Recombination hyb.A-BS Recombination hyb.BAB Gene conversion hyb.B-A Recombination ex3 EET ex4 GETGQL ex3 EET ex4 GEMGQL ex3 EET ex4 GETGQL ex2 QTN ex4 GERVQL HIL SJL HIL Sta Johe,Vengelen-Tyler,Leger,Blumenfeld Blood 1991,78:2456 Examples were provided showing that, on the red cell surface, single amino acid or carbohydrate alterations, resulting from missense mutations or other DNA changes are recognized, as one might expect, as foreign by the immune system and, remarkably, can be detected by serological approaches . Acknowledgements Contributors to the database Departments of Biochemistry and Cell Biology, Albert Einstein College of Medicine Thank you!