* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download 401Lecture8Sp2013post
Survey
Document related concepts
Transcript
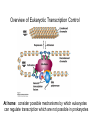
Outline • • • • • • Molecular Cell Biology Assessment Review from last lecture Role of nucleoporins in transcription Activators and Repressors Epigenetic mechanisms of inheritance Cell type specification – Restriction of cell fates – Embryonic stem cells • Pluripotency • Chromatin structure On-line assessment • • • • • • Molecular and cellular biology test Multiple true/false format Multi-institution study 3 points for participation Link will be emailed today Complete by next Tuesday (6/4) Conclusions from Pax6 Gene • Different control regions can control transcription of the same gene in different cell types • Different subsets of transcription factors bind to control regions of the same gene in different cell types • Control regions can be located far from transcription start sites From Last Lecture: Techniques to Characterize CisActing Regulatory Regions • Transgenic mice – Create DNA constructs containing potential cis-acting sequences + reporter gene and introduce into embryonic stem cells • Reporter Gene Assay (fig. 7-25) – Cell culture approach • Linker Scanning Mutation Analysis (fig. 7-21) – In vivo or in cell culture • DNase I Footprinting (fig. 7-23) • Comparative genomics Purple = in text book readings From Last Lecture: Linker-scanning Mutation of a Gene Regulatory Region Reporter gene activity What can you tell from comparing mutants 1, 2, 3 & 4? Fig. 7-21 Review at home: Techniques to Identify Regulatory Elements Comparison of Eukaryotic Control Regions Structure of a Transcription Factor What do you notice when you compare (a) and (c)? What do you notice when you compare (a) and (b)? Fig. 7-22. Lodish Example of a Typical Mammalian Enhancer Enhanceosome on the β-interferon enhancer What three things determine whether a given protein is found associated with the enhancer? • WT1 represses EGR-1 • SRF/TCF and AP1 are transcription activators • Based on this schematic, how is the mechanism of action different for the eukaryotic WT1 (Wilms Tumor) repressor compared to prokaryotic repressors (e.g. lac i)? Summary • Activators promote transcription and are modular proteins composed of a DNA binding domain and an activation domain • Repressors inhibit transcription and are modular proteins composed of a DNA binding domain and a repressor domain • Both repressor and activators recruit other proteins to affect gene expression • A cell must produce the specific set of activators required for transcription of a particular gene in order to express that gene Can Proteins Serve Both a Regulatory and a Structural Function in the Cell? * = Dynamic D’Angelo et al. 2009 Not discussed – you are not responsible for this information A. Chromatin IP experiment identifying binding sites for Nup98 and lamin What do you notice about the binding sites for Nup 98 and Lamin on this chromosome? What does this suggest about the location of Nup98 in the cell? Capelson et al., Cell 140: 372 (2010) Not discussed – you are not responsible for this information What does the expression data tell us about the genes that Nup98 associates with? What do these data suggest about the role(s) of Nup98? What else are dynamic nucleoporins doing? B. Expression levels of Nup98-associated genes Capelson et al., Cell 140: 372 (2010) Overview of Eukaryotic Transcription Control At home: consider possible mechanisms by which eukaryotes can regulate transcription which are not possible in prokaryotes How are established expression patterns inherited? Cell Memory Epigenetic mechanisms of inheritance – DNA methylation (DNA methyltransferases) – Histone modifications (e.g. methylation) – noncoding RNAs Memory of Winter PRC2/VRN2= Polycomb Group Complex (PcG) Novel mechanisms for Gene Repression: Epigenetic Silencing of the Flowering Gene FLC by a Noncoding RNA Luciferase activity COLDAIR transcription is induced by cold COLDAIR noncoding RNA binds Polycomb group proteins FLC locus is methylated on histone H3 Silencing Science 331: 76-79 (2011) Cascade of Gene Expression Required for Vernalization-Induced Silencing Science 331: 76-79 (2011) Strategy for Epigenetic Inheritance of Histone Modifications Reader/Writer Hypothesis How is Histone H3 methylation Maintained during DNA replication What two properties Must the H3K9 HMT have? Fig. 7-46. Lodish et al. 2013 Model for Repression by Polycomb Complexes: Establishing Gene Repression Fig. 7-47 PRC2 contains a histone methyl transferase (writer) Where have we seen PRC2 before? What is the role of the repressor? Model for Repression by Polycomb Complexes: Maintaining gene Repression PRC1(the reader) contains a protein (Pc) that binds H3K27Me Not shown: PRC2 (the writer) can bind PRC1 to maintain the methyl mark after DNA replication Do plants have PRC1 complexes? Fig. 7-47 Conclusions • Changes in chromatin structure are necessary before transcription can occur at most genes • Activators can direct histone acetylation and methylation to specific genes • Repressors can direct histone deacetylation, histone methylation and DNA methylation to specific genes • Repressors and microRNAs can recruit polycomb group complexes to promote heterochromatin formation • Epigenetic marks can be inherited by daughter cells