* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Protein Structure
Gene expression wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Expression vector wikipedia , lookup
Magnesium transporter wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Interactome wikipedia , lookup
Point mutation wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Protein purification wikipedia , lookup
Western blot wikipedia , lookup
Homology modeling wikipedia , lookup
Genetic code wikipedia , lookup
Metalloprotein wikipedia , lookup
Peptide synthesis wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Biosynthesis wikipedia , lookup
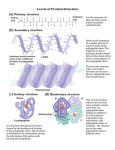
Protein Structure Dr Vivek Joshi, MD Peptides and Proteins Peptide: A short polymer of amino acids joined by peptide bonds; they are classified by the number of amino acids in the chain. Dipeptide: A molecule containing two amino acids joined by a peptide bond. Tripeptide: A molecule containing three amino acids joined by peptide bonds. Polypeptide: A macromolecule containing many amino acids joined by peptide bonds. Protein: A biological macromolecule containing at least 30 to 50 amino acids joined by peptide bonds. 2 2 Levels of Organization Protein Structure Primary structure: The sequence of amino acids in a polypeptide chain; read from the N-terminal amino acid to the C-terminal amino acid. Secondary structure: Conformations of amino acids in localized regions of a polypeptide chain; examples are ahelix, b-pleated sheet, and random coil. Tertiary structure: The complete three-dimensional arrangement of amino acids in a polypeptide chain. Quaternary structure: The spatial relationship and interactions between subunits in a protein that has more than one polypeptide chain. 3 3 Levels of Organization in Protein Structure (contd.) 4 4 Four levels of protein structure Primary Structure ◦ Order in which amino acids are joined together ◦ AA sequence of the polypeptide chain ◦ Polypeptide backbone ◦ Includes the location of any disulfide bond ◦ Higher levels of protein structure are decided by primary structure ◦ Amino acid sequence is specific –Coded by gene ◦ Determines ‘Biological activity’ of the protein Stabilized by covalent bonds Peptide Bond Formation Peptide bond – Amide linkage between the carboxyl group of one amino acid and the a-amino group of another A condensation reaction with liberation of water Characteristics of the peptide bond Shows Electronic resonance Has a partial double bond character No freedom of rotation Rigid and planar-All 6 of the atoms O, C, N, H are coplanar Characteristics of the Peptide Bond Generally a trans bond-Because of the steric interference of the R-groups when in the cis position Uncharged but polar Naming Peptides Free amino end of the peptide chain (N-terminal) is written to the left Free carboxyl end of the peptide chain (C-terminal) is written to the right All amino acid sequences are read from the N- to the C-terminal end of the peptide Tyrosylglycylglycylphenylalanylleucine Primary Structure Amino acid sequence specifies 3D structure The 20 amino acids can be linked in combinations specific for a given protein Long chains of amino acids fold in a pattern dependent on the exact order of amino acids Different regions of the polypeptide chain may assume different conformations as determined by the sequence of amino acids in that region A genetic mutation leading to one incorrect amino acid substitution in a protein comprising thousands of amino acids can result in that protein having a different shape and little or even no biological activity-sickle cell disease . Secondary Structure Defines the steric relationship between amino acids that are close to each other in the primary amino acid sequence Brought about by linking the carbonyl and amide groups of the peptide bonds by means of hydrogen bonds (H-bonds) Folding of polypeptide chain along a single axis Kinds: Alpha helix Beta sheets Beta turns Secondary Structure Alpha helix A rigid, rod-like structure The lowest energy and most stable conformation for a polypeptide chain Forms spontaneously Peptide bond planes are parallel to the axis of the helix Stability arises from the formation of the maximum possible number of H-bonds 3.6 amino acid residues per 360° turn (5.4 Å) 13 atoms/turn 1.5 Å rise/residue R groups extend outward Right-handed Alpha helix ◦ H-bonds are formed between Each carbonyl oxygen atom of a peptide bond and the hydrogen attached to the amide nitrogen of the peptide bond 3.6 amino acid residues further along the polypeptide chain Each peptide bond forms 2 H-bonds: One to the peptide bond of the 4th residue above One to the peptide bond of the 4th residue below Destabilized by the presence of proline An helix breaker Interrupts a-helical structure because Its ring structure exerts geometric constraint The nitrogen in the peptide linkage does not contain the H atom required to form H-bonds Alpha helix 15 Secondary Structure Beta pleated sheet Composed of 2 or more peptide chains or segments of polypeptide chains that are arranged either parallel or anti-parallel to each other C N N C Antiparallel N C N C Parallel Secondary Structure Beta pleated sheet the α-carbon and its R group side chain alternate slightly above and slightly below the plane of the main chain of the polypeptide (ruffled or pleated appearance) Secondary Structure Beta bend (reverse turn) ◦ ◦ ◦ ◦ ◦ Makes a tight 180° turn Allows the polypeptide chain to abruptly reverse its direction Helps to form a compact, globular shape Surface of proteins-Charged residues Generally composed of : Glycine – has the smallest R group Proline – causes a kink in the polypeptide chain Combinations of secondary structures Form the interior (core region) of globular proteins Connected by loop regions at the surface of the protein Zn finger motif-DNA bab Hairpin aa Supersecondary Structures (Motifs) Tertiary structure 3-dimensional arrangement of all atoms in a single polypeptide chain The entire protein molecule coils into an overall threedimensional shape-Functional property to the protein Spatial arrangement of amino acid residues that are far apart in a linear sequence Superfolding brings functional groups that are far apart near to each other. Correct folding is assisted by Chaperons Incorrect folding-Altered protein-Prion disease Tertiary Structure Stabilizing interactions: Covalent: disulfide bonds Non-covalent Hydrogen bonds. Fine tunes the tertiary structure by selecting the unique native structure of a protein Ionic interactions. The association of two ionic protein groups of opposite charge is known as ion pair or salt bridge. Hydrophobic interactions- The major determinant of protein native structure The conformation or shape of the protein is determined by: ◦ The nature of the interaction of the different side chains with the aqueous environment ◦ The interactions of the different side chains with the other side chains Quaternary Structure Refers to the spatial arrangement of the polypeptide chains of a multi-chain protein(2 />2 polypeptide chains) and the nature of their contacts Each polypeptide chain is a monomer/ subunit Dimer, tetramer, Oligomeric protein - protein with multiple subunits Homooligomers – have identical subunits Heterooligomers – have several distinct polypeptide chains Non-covalent interactions hold the subunits together Hydrophobic, ionic, H-bond Hemoglobin-2 a ,2 b Immunoglobulin-2 Heavy ,2 Light chain Creatine Kinase-Dimer Lactate Dehydrogenase-Tetramer Quaternary structure refers to the interaction among protein subunits Hemoglobin contains 2aand 2b subunits Subunits may function independently or may work cooperatively Hemoglobin – binding of oxygen to 1 subunit of the tetramer increases the affinity of the other subunits to oxygen-Cooperative binding Denaturation of Proteins Denaturation: The process of destroying the native conformation of a protein by chemical or physical means. ◦ Usually denaturation of protein refers to disruption of tertiary and secondary structure, while primary structure remain unaffected. ◦ It leads to loss of biological activity ◦ It decreases solubility and increases precipitability ◦ Some denaturations are reversible, while others permanently damage the protein. Denaturing agents include: ◦ Heat, 6 M urea, detergents, Acids, Bases, Salts, Reducing agents, Heavy metals, Alcohol 25 25 Protein Turnover Older proteins break down-Replaced by new one. Protein synthesis –Translation-Ribosomes Protein Breakdown Lysosomes-Proteases-Endocytosed protein Proteasomes-Cytoplasmic complexes-Older and abnormal protein Proteasomes are large protein complexes inside all eukaryotes and in some bacteria. In eukaryotes, they are located in the nucleus and the cytoplasm. The main function of the proteasome is to degrade unneeded or damaged proteins by proteolysis, a chemical reaction that breaks peptide bonds. 26 Proteasome-Ubiquitin pathway Proteins are tagged for degradation by a small protein called ubiquitin. The tagging reaction is catalyzed by enzymes called ubiquitin ligases. Once a protein is tagged with a single ubiquitin molecule, this is a signal to other ligases to attach additional ubiquitin molecules. The result is a polyubiquitin chain that is bound by the proteasome, allowing it to degrade the tagged protein. Protein Misfolding Misfolded protein-Tagged and degraded within the cell. Defect in this control system-Extracellular and intracellular accumulation of misfolded proteins Age Disease Prion Disease Amyloidosis 28 Prion Disease Prion diseases are a family of degenerative brain disorders observed in humans and numerous other mammals. Some (but not all) cases originate by transmission from one individual to another, however, no bacterial, viral, or parasitic agent has been identified Propagate by transmitting mis-folded protein Protein does not itself self-replicate Process dependent on presence of polypeptide in the host Protein conformational disease Transmitted by altering the conformation Creutzfeldt-Jakob disease (CJD) in humans. Prion-related Protein (PrP) Encoded by the PrP gene of the mammal Exists in 2 isoforms -identical primary structures and posttranslational modifications -different tertiary and quaternary structures #PrPc – PrP-sen (proteinase-sensitive) Normal cellular protein On the outer surface of neurons Function still unclear #PrPSc – PrP-res (proteinase-resistant) Pathologic isoform Invariably associated with TSEs or prion diseases Prion-related Protein (PrP) PrPc – Rich in a-helix PrPSc – Consists mostly of b-sheets Conversion of PrPc into PrPSc Involves alteration of ahelical structure into b-sheet-Aggregate-Amyloid Spontaneous Caused by infection Consumption of food, especially neural tissue, that contains PrPSc Neurodegenerative diseases -Alzheimer's, Parkinson's, Huntington's Neuronal dysfunction - Induced by diffusible oligomers of misfolded proteins. What bonds are broken and remade to go from a-helix to b-sheets? Prion-related Protein (PrP) Prions (Proteinaceous infectious particle) Infectious proteins that contain no nucleic acid Protein itself is capable of acting like infectious material and modifying protein content of the entire organism Cause transmissible spongiform encephalopathies (TSE) or prion diseases Amyloidosis Normal Amyloid precursor protein-Abnormal proteolytic cleavage-Long Fibrillar protein consisting of β-pleated sheetAmyloids Amyloid in Alzheimer’s disease –Amyloid β(A β)Neurotoxic Thank You