* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Slides - Department of Computer Science • NJIT
Gel electrophoresis of nucleic acids wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Gene expression wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Bisulfite sequencing wikipedia , lookup
Community fingerprinting wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Molecular cloning wikipedia , lookup
DNA supercoil wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Proteolysis wikipedia , lookup
Genetic code wikipedia , lookup
Non-coding DNA wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Point mutation wikipedia , lookup
Biosynthesis wikipedia , lookup
Lecture 1
BNFO 240
Usman Roshan
Course overview
• Perl progamming language (and some Unix basics)
• Sequence alignment problem
– Algorithm for exact pairwise alignment
– Heuristics for exact multiple alignment
– Computational complexity
– Heuristics for pairwise alignment and BLAST, FASTA database
search
– Real world alignment problems
– Substitution matrices
• Phylogeny reconstruction
– Estimating distance matrices
– Distance based phylogeny reconstruction ---- UPGMA and
neighbor joining algorithms
Overview (contd)
• Wednesdays --- meet in GITC 2305
• Fridays --- meet in PC Mall room number
PC 36
• Grade: 50% monthly programming
assignment and 50% final exam
• Texts:
– Introduction to Bioinformatics by Arthur Lesk
– Beginning Perl for Bioinformatics by James
Tisdall
DNA Sequence Evolution
-3 mil yrs
AAGACTT
AAGACTT
AAGGCTT
AAGGCTT
_GGGCTT
_GGGCTT
GGCTT
_G_GCTT
(Mouse)
(Mouse)
TAGACCTT
TAGACCTT
TAGGCCTT
TAGGCCTT
(Human)
(Human)
-2 mil yrs
T_GACTT
T_GACTT
TAGCCCTTA
TAGCCCTTA
(Monkey)
(Monkey)
A_CACTT
A_CACTT
ACACTTC
A_CACTTC
(Lion)
ACCTT
A_C_CTT
(Cat)
(Cat)
-1 mil yrs
today
Comparative bioinformatics
• What is the evolutionary relationship of a
set of DNA sequences?
• What are the evolutionary conserved
regions of a set of proteins?
• How evolutionary close is a pair of
species?
• How similar are two DNA sequences?
• How similar are a set of DNA sequences?
Representing DNA in a format
manipulatable by computers
• DNA is a double-helix molecule made
up of four nucleotides:
–
–
–
–
Adenosine (A)
Cytosine (C)
Thymine (T)
Guanine (G)
• Since A (adenosine) always pairs with
T (thymine) and C (cytosine) always
pairs with G (guanine) knowing only
one side of the ladder is enough
• We represent DNA as a sequence of
letters where each letter could be
A,C,G, or T.
• For example, for the helix shown here
we would represent this as CAGT.
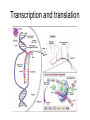
Transcription and translation
Amino acids
Proteins are chains of
amino acids. There are
twenty different amino
acids that chain in
different ways to form
different proteins.
For example,
FLLVALCCRFGH
(this is how we could store
it in a file)
This sequence of amino
acids folds to form a 3-D
structure
Protein folding
Protein folding
• The protein folding
problem is to determine
the 3-D protein structure
from the sequence.
• Experimental techniques
are very expensive.
• Computational are cheap
but difficult to solve.
• By comparing sequences
we can deduce the
evolutionary conserved
portions which are also
functional (most of the time).
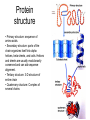
Protein
structure
• Primary structure: sequence of
amino acids.
• Secondary structure: parts of the
chain organizes itself into alpha
helices, beta sheets, and coils. Helices
and sheets are usually evolutionarily
conserved and can aid sequence
alignment.
• Tertiary structure: 3-D structure of
entire chain
• Quaternary structure: Complex of
several chains
Key points
• DNA can be represented as strings
consisting of four letters: A, C, G, and T.
They could be very long, e.g. thousands
and even millions of letters
• Proteins are also represented as strings
of 20 letters (each letter is an amino acid).
Their 3-D structure determines the
function to a large extent.
Pairwise sequence alignment
• How to align two sequences?
Pairwise alignment
• How to align two sequences?
• We use dynamic programming
• Treat DNA sequences as strings over the
alphabet {A, C, G, T}
Pairwise alignment
Dynamic programming
Define V(i,j) to be the optimal pairwise alignment
score between S1..i and T1..j (|S|=m, |T|=n)
Dynamic programming
Define V(i,j) to be the optimal pairwise alignment
score between S1..i and T1..j (|S|=m, |T|=n)
Time and space complexity is O(mn)
Tabular computation of scores
Traceback to get alignment
How do we understand this dynamic
programming algorithm?
• Let’s first look at some example
alignments
• Let’s look at gaps. How do we know where
to insert gaps
• Let’s look at the structure of an optimal
alignment of two sequences x and y and
how it relates optimal alignments of
subsequences of x and y