* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download ppt
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Genetic code wikipedia , lookup
Oxidative phosphorylation wikipedia , lookup
Evolution of metal ions in biological systems wikipedia , lookup
Adenosine triphosphate wikipedia , lookup
Peptide synthesis wikipedia , lookup
Fatty acid synthesis wikipedia , lookup
Fatty acid metabolism wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Proteolysis wikipedia , lookup
Oligonucleotide synthesis wikipedia , lookup
Biochemistry wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Citric acid cycle wikipedia , lookup
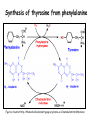
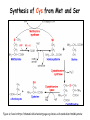
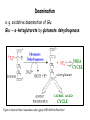
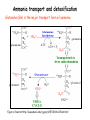
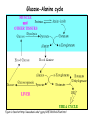
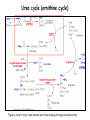
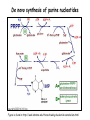
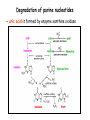
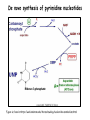
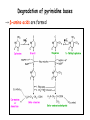
Metabolism of amino acids, purine and pyrimidine bases Pavla Balínová Amino acids (AAs) Sources of AAs: • diet • synthesis de novo • protein degradation dietary proteins body proteins proteosynthesis AAs pool de novo biosynthesis (E,glc,fat) N-compound synthes. degradation Biosynthesis of amino acids (AA) Humans can synthesize only 10 of the 20 AA. Essential AA = AA that cannot be synthesized „de novo“. They must be obtained from diet. Nonessential AA: Ala is synthesized from pyruvate. Cys is synthesized from Met and Ser. Tyr is formed by hydroxylation from Phe. Essential AA Nonessential AA Arg Ala His Asn Ile Asp Leu Cys Lys Gln Met Glu Phe Gly Thr Pro Trp Ser Val Tyr Synthesis of AAs in a human body - 5 substrates 1. oxaloacetate → Asp, Asn 2. -ketoglutarate → Glu, Gln, Pro, (Arg) 3. pyruvate → Ala 4. 3-phosphoglycerate → Ser, Cys, Gly 5. Phe → Tyr Synthesis of thyrosine from phenylalanine Figure is found at http://themedicalbiochemistrypage.org/amino-acid-metabolism.html#tyrosine Formation of activated methionine = S-adenosylmethionine (SAM) SAM is used as –CH3 group donor in metabolic methylations Figure is found at http://themedicalbiochemistrypage.org/amino-acid-metabolism.html#cysteine Synthesis of Cys from Met and Ser Figure is found at http://themedicalbiochemistrypage.org/amino-acid-metabolism.html#cysteine Degradation of AA 20 different multienzyme sequences exist for catabolism of AAs. All common 20 AAs are converted to only 7 compounds: • pyruvate • acetyl-CoA • acetoacetyl-CoA • α-ketoglutarate • succinyl-CoA • fumarate • oxaloacetate Three types of reactions are typical for degradation of AAs: 1. Transamination 2. Deamination 3. Decarboxylation Transamination = an exchange of –NH2 between amino acid and α-ketoacid These reactions are catalyzed by transaminases (aminotransferases). Most of them require α-ketoglutarate as an acceptor of –NH2. Coenzyme of transaminases: pyridoxal phosphate (vit. B6 derivative) Figure is found at http://web.indstate.edu/thcme/mwking/nitrogen-metabolism.html Aminotransferases (transaminases) important in medicine Alanine aminotransferase (ALT) Aspartate aminotransferase (AST) Figure was adopted from Devlin, T. M. (editor): Textbook of Biochemistry with Clinical Correlations, 4th ed. Wiley-Liss, Inc., New York, 1997. ISBN 0-471-15451-2 Deamination e. g. oxidative deamination of Glu Glu → α-ketoglutarate by glutamate dehydrogenase Figure is found at http://www.sbuniv.edu/~ggray/CHE3364/b1c25out.html Decarboxylation → primary amines a) decarboxylation of His → histamine b) decarboxylation of Trp → serotonin c) decarboxylation of Tyr → epinephrine and norepinephrine d) decarboxylation of Glu → GABA (γ-aminobutyrate) Figure is found at http://www.sbuniv.edu/~ggray/CHE3364/b1c25out.html Ammonia transport and detoxification Glutamine (Gln) is the major transport form of ammonia. Figure is found at http://www.sbuniv.edu/~ggray/CHE3364/b1c25out.html Glucose-Alanine cycle Figure is found at http://www.sbuniv.edu/~ggray/CHE3364/b1c25out.html Urea cycle (ornithine cycle) • • • • • • substrates: NH4+, HCO3-, Asp, ATP product: urea function: synthesis of non-toxic urea subcellular location: mitochondria and cytosol organ location: liver regulatory enzyme: carbamoyl phosphate synthetase I Urea cycle (ornithine cycle) l Figure is found at http://web.indstate.edu/thcme/mwking/nitrogen-metabolism.html The fate of carbon skeletons of AA during catabolism • The strategy of the cell is to convert carbon skeletons to compounds useful in gluconeogenesis or CAC. • Glucogenic AAs = AA that can form any of intermediates of carbohydrate metabolism Gly, Ala, Ser, Cys, Thr → pyruvate Glu, Pro, Arg, His → Glu → α-ketoglutarate Met, Ile, Val → succinyl-CoA • Ketogenic AAs are converted to acetyl-CoA and acetoacetyl-CoA. They yield ketone bodies. Leu, Lys ● Glucogenic-ketogenic AAs = Thr, Phe, Tyr, Ile Figure is found at http://www.biocarta.com/pathfiles/glucogenicPathway.asp F De novo synthesis of purine nucleotides Figure is found at http://web.indstate.edu/thcme/mwking/nucleotide-metabolism.html Important notes about biosynthesis of purine nucleotides • Subcellular location: cytoplasm • PRPP = phosphoribosyl pyrophosphate is derived from ribose-5-P • IMP = inosine monophosphate serves as the common precursor of AMP and GMP synthesis • Gln, Gly, Asp are donors of C and N atoms • CO2 is a source of C • C1 units are transferred via tetrahydrofolate „Salvage pathway“: • purines from normal turnover of cellular NA can be converted to nucleoside triphosphates • substrates: purine bases, PRPP, ATP Degradation of purine nucleotides → uric acid is formed by enzyme xanthine oxidase De novo synthesis of pyrimidine nucleotides Figure is found at http://web.indstate.edu/thcme/mwking/nucleotide-metabolism.html Important notes about synthesis of pyrimidine nucleotides • Carbamoyl phosphate is formed from Gln and CO2 (2 ATP are consumed). This reaction is catalyzed by carbamoyl-P synthetase II (cytosolic enzyme) = regulatory step • pathway occurs in cytoplasm but formation of orotate occurs in mitochondrion → orotate is linked by PRPP → OMP → UMP • UMP → UTP → CTP TTP TTP ● UTP inhibits regulatory enzyme, activator is PRPP „Salvage pathway“: ● pyrimidine nucleosides are phosphorylated (ATP) to nucleotides Degradation of pyrimidine bases → β-amino acids are formed