* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Viral Structure Lec. 2
Survey
Document related concepts
Homology modeling wikipedia , lookup
Structural alignment wikipedia , lookup
Protein folding wikipedia , lookup
Bimolecular fluorescence complementation wikipedia , lookup
Circular dichroism wikipedia , lookup
Protein domain wikipedia , lookup
Protein purification wikipedia , lookup
Protein structure prediction wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
List of types of proteins wikipedia , lookup
Protein mass spectrometry wikipedia , lookup
Western blot wikipedia , lookup
Transcript
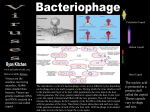
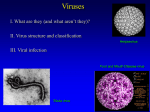
Viral Structure Structure of a Virus • Questions Relating to Structure – Is it rigid? – How big is it? – Is it flexible • Structure Must Serve Virus – It should provide protection for genome – It should allow virus to move from one host to next – It should allow for attachment of virus on to new host Structural Components • Viral Capsid – Capsid means “box” • Capsid can be square, tetrahedron or icosahedron – Capsid is made up of protein – Capsid is the storage site for genome – Many capsids have a ‘shell’ structure – Genome + Capsid = Nucleocapsid – Capsid is made up of polymeric proteins to conserve genome • Ex. 5 Kb genome requires 30,000 a/a capsid, which means 90 Kb genome just for capsid!! • Solution: use multiple copies of same protein • Viral Envelope – It is the covering of the nucleocapsid – Made up of a phospholipid bilayer – It should allow for attachment of virus on to new host Tools for Studying Viral Structure • Electron Microscopy – Excellent tool with some limitations • High resolution • Image can be a distortion due to specimen processing • X-ray Diffraction – Good for naked virions (no envelope) • Cryoelectron Microscopy – Flash frozen with liquid nitrogen Structural Symmetries • Icosahedral Symmetry – 20 triangular faces – It is a common capsid structure – Examples of viruses with icosahedral symmetry • Parvoviruses – – – – – These are simple viruses 5 Kb ssDNA genome Capsid is formed with 60 copies of single protein Protein is approximately 520 a/a 1/3 of genome is dedicated to capsid • Polio virus – Uses 180 copies of 3 subunit proteins – Much bigger virus •Desmodium Yellow Mottle Virus •X-Ray Crystallography •Icosahedral Symmetry Other Structural Symmetries Fig. 2.7 The helical nucleocapsid of tobacco mosaic virus. Other Structural Proteins • Core Proteins – Can originate from host • Ex. Histones – Can also be virally coded – Their function is to condense viral genome • Scaffolding Proteins – Facilitate capsid formation – Facilitate docking of proteins – Facilitate stability of proteins during assembly – Not included inside virion Viral Envelope • • • • Lipid bilayer Most originate from cellular host Cholesterol and glycoproteins are present In cases where budding occurs at the plasma membrane (Ex. Influenza) envelope resembles host’s plasma membrane i.e cholesterol and phospholipids • In cases where budding occurs at the ER (Ex. Flaviviruses) envelope has less cholesterol, similar to ER Viral Glycoproteins • Glycoproteins – Short cytoplasmic tail – Hydrophobic segment for anchoring (~20 amino acids) – Relatively large ectodomain (external domain) • Ectodomain – Extensively glycosylated preventing aggregation of virions – Glycosylation attracts water and reduces sticking (carried out in ER) – Palmitoylation of cysteine residues is also extensive (carried out in ER) • Most envelope proteins are type I – That means N-terminus facing out, C terminus near anchor domain – Some though are type II Viral Budding • Several budding mechanims exist • Envelope proteins create 2-D pools excluding cellular memberane proteins • Capsids can bind ‘cytoplasmic’ tails of envelope proteins – Binding of capsid proteins and envelope proteins is mediated by matrix proteins in viruses with helical nucleocapsids (Ex. influenza budding) • When budding takes place eventually, capsid ‘wraps’ itself around host plasma membrane Influenza Viral Budding Matrix protein (M) interacts with HA and NA HA are glycoproteins on envelope Interaction occurs at the level of their cytoplasmic tail M protein also interacts with helical nucleocapsid proteins RNP Semliki Forrest Viral Budding Direct interaction between icosahedral capsid and envelope proteins No Matrix protein involved Interaction at the level of cytoplasmic tail of envelope protein