* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Lecture 27
Evolution of metal ions in biological systems wikipedia , lookup
Metabolic network modelling wikipedia , lookup
Photosynthetic reaction centre wikipedia , lookup
Oxidative phosphorylation wikipedia , lookup
Fatty acid metabolism wikipedia , lookup
Adenosine triphosphate wikipedia , lookup
Proteolysis wikipedia , lookup
Nitrogen cycle wikipedia , lookup
Genetic code wikipedia , lookup
Epitranscriptome wikipedia , lookup
Metalloprotein wikipedia , lookup
Biochemistry wikipedia , lookup
Citric acid cycle wikipedia , lookup
Amino acid synthesis wikipedia , lookup
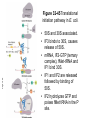
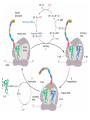
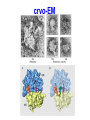
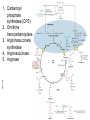
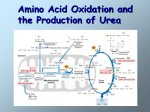
FCH 532 Lecture 21 Chapter 32: Translation Quiz today (Wed.) on amino acids Quiz on Friday TCA cycle Quiz on Monday Transamination mechanism. Page 987 Page 1321 Figure 32-43 Some translational initiation (ShineDalgarno) sequences recognized by E. coli ribosomes. Shine-Dalgarno sequences typically start 10-15 nt upstream of the initiation codon. Are only found in prokaryotes. Page 1323 Figure 32-45 Translational initiation pathway in E. coli. • 50S and 30S associated. • IF3 binds to 30S, causes release of 50S. • mRNA, IF2-GTP (ternary complex), fMet-tRNA and IF1 bind 30S. • IF1 and IF2 are released followed by binding of 50S. • IF2 hydrolyzes GTP and poises fMet tRNA in the P site. Page 1327 Defining tRNA Binding Sites in Different functional States GENERATE RIBOSOMES IN THE FOLLOWING STATES: A (Aminoacyl): EF-Tu.GTP dependent; mRNA dependent; occupied P Site P (Peptidyl): Reactive with Puromycin (Pm) E (Exit): Deacylated tRNA MONITOR BY CHEMICAL FOOTPRINTING: 30S A site protections: 50S P site protections (also X-linkers, EDTA-FeII) Looking at footprint pre and post peptide bond, translocation The data didn't fit into a simple 2 site model HYBRID STATES HAD TO BE INVOKED tRNA movement occurs independently on 2 subunits via 6 hybrid states. 1. A/T --> 2. A/A --> 3. A/P --> 4. P/P --> 5. P/E --> 6. E In this model the tRNA would "ratchet" its way through the ribosome undergoing 50° rotations along its longitudinal axis from A to P. This model has received support from EM and X-ray studies. cryo-EM Aminoacyl-tRNA EF-Tu-GTP EF-Ts GTP EF-Tu-EF-Ts Page 1333 EF-Ts GDP RF-1 = UAA RF-2 = UAA and UGA Cannot bind if EF-G is present. RF-3-GTP binds to RF1 after the release of the polypeptide. Hydrolysis of GTP on RF-3 facilitates the release of RF-1 (or RF-2). Page 1335 EF-G-GTP and ribosomal recycling factor (RRF)-bind to A site. Release of GDP-RF-3 EF-G hydrolyzes GTP -RRF moves to the P site to displace the tRNA. RRF and EF-G-GDP are released yielding inactive 70S Translation • • • • Shine-Dalgarno sequence Initiation Elongation Release Page 1322 Page 1336 Figure 32-61 Ribosome-catalyzed hydrolysis of peptidyl–tRNA to form a polypeptide and free tRNA. Urea Cycle • • • • • • • Excess nitrogen is excreted after the metabolic breakdown of amino acids in one of three forms: Aquatic animals are ammonotelic (release NH3 directly). If water is less plentiful, NH3 is converted to less toxic products, urea and uric acid. Terrestrial vertebrates are ureotelic (excrete urea) Birds and reptiles are uricotelic (excrete uric acid) Urea is made by enzymes urea cycle in the liver. The overall reaction is: NH3+ NH3 + HCO3- + -OOC-CH2-CH-COOAsp O 3ATP 2ADP + 2Pi + AMP + PPi NH2-C-NH2 + -OOC-CH=CH-COOUrea Fumarate Urea Cycle • • • 2 urea nitrogen atoms come from ammonia and aspartate. Carbon atom comes from bicarbonate. 5 enzymatic reactions used, 2 in the mitochondria and 3 in the cytosol. NH3+ NH3 + HCO3- + -OOC-CH2-CH-COOAsp O 3ATP 2ADP + 2Pi + AMP + PPi NH2-C-NH2 + -OOC-CH=CH-COOUrea Fumarate Page 992 Carbamoyl phosphate synthetase • • Carbamoyl phosphate synthetase (CPS) catalyzes the condensation and activation NH3 and HCO3- to form carbomyl phosphate (first nitrogen containing substrate). Uses 2 ATPs. O 2ATP + NH3 + HCO3- NH2-C-OPO3- + 2ADP + 2Pi Carbamoyl phosphate • 1. 2. Eukaryotes have 2 types of CPS enzymes Mitochondrial CPSI uses NH3 as its nitrogen donor and participates in urea biosynthesis. Cytosolic CPSII uses glutamine as its nitrogen donor and is involved in pyrimidine biosynthesis. Figure 26-8 The mechanism of action of CPS I. • 1. Page 993 CPSI reaction has 3 steps Activation of HCO3- by ATP to form carboxyphosphate and ADP. 2. Nucelophilic attack of NH3 on carboxyphosphate, displacing the phsophate to form carbamate and Pi. 3. Phosphorylation of carbamate by the second ATP to form carbamoyl phosphate and ADP The reaction is irreversible. Allosterically activated by Nacetylglutamate. Figure 26-9 X-Ray structure of E. coli carbamoyl phosphate synthetase (CPS). • • • • Page 993 • E. coli has only one CPS (homology to CPS I and CPS II) Heterodimer (inactive). Allosterically activated by ornithine (heterotetramer of (4). Small subunit hydrolyzes Gln and delivers NH3 to large subunit. Channels intermediate of two reactions from one active site to the other. Page 992 Ornithine transcarbomylase • • • • Transfers the carbomoyl group of carbomyl phosphate to ornithine to make citrulline Reaction occurs in mitochondrion. Ornithine produced in the cytosol enters via a specific transport system. Citrulline is exported from the mitochondria. Page 992 Arginocuccinate Synthetase • • • • 2nd N in urea is incorporated in the 3rd reaction of the urea cycle. Condensation reaction with citrulline’s ureido group with an Asp amino group catalyzed by arginosuccinate synthetase. Ureido oxygen is activated as a leaving group through the formation of a citrulyl-AMP intermediate. This is displaced by the Asp amino group to form arginosuccinate. Page 994 Figure 26-10 The mechanism of action of argininosuccinate synthetase. Arigininosuccinase and Arginase • • • • • • Argininosuccinse catalyzes the elimination of Arg from the the Asp carbon skeleton to form fumurate. Arginine is the immediate precursor to urea. Fumurate is converted by fumarase and malate dehydrogenase to to form OAA for gluconeogenesis. Arginase catalyzes the fifth and final reaction of the urea cycle. Arginine is hydrolyzed to form urea and regenerate ornithine. Ornithine is returned to the mitochondria. Page 992 1. Carbamoyl phosphate synthetase (CPS) 2. Ornithine transcarbamoylase 3. Argininosuccinate synthetase 4. Arginosuccinase 5. Arginase Regulation of the urea cycle • • • • • Carbamoyl phosphate synthetase I is allosterically activated by N-acetylglutamate. N-acetylglutamate is synthesized from glutamate and acetylCoA by N-acetylglutamate synthase, it is hydrolyzed by a specific hydrolase. Rate of urea production is dependent on [N-acetylglutamate]. When aa breakdown rates increase, excess nitrogen must be excreted. This results in increase in Glu through transamination reactions. Excess Glu causes an increase in N-acetylglutamate which stimulates CPS I causing increases in urea cycle. Metabolic breakdown of amino acids • • • • Degradation of amino acids converts the to TCA cycle intermediates or precursors to be metabolized to CO2, H2O, or for use in gluconeogenesis. Aminoacids are glucogenic, ketogenic or both. Glucogenic amino acids-carbon skeletons are broken down to pyruvate, -ketoglutarate, succinyl-CoA, fumarate, or oxaloacetate (glucose precursors). Ketogenic amino acids, are broken down to acetyl-CoA or acetoacetate and therefore can be converted to fatty acids or ketone bodies.