* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Document
Chemical weapon wikipedia , lookup
Peptide synthesis wikipedia , lookup
Interactome wikipedia , lookup
Genetic code wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Drug discovery wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Protein structure prediction wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Biosynthesis wikipedia , lookup
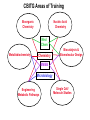
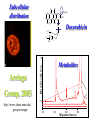
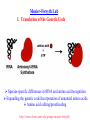
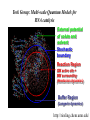
Chemical Biology Specialty Area at U of M Our program emphasizes highly interdisciplinary research and training, aimed at the development and integration of modern chemical methods in order to understand biological problems at the molecular level. Chemical Biology Specialty Area at U of M 19 Faculty Highly Interdisciplinary Research NIH Chemistry and Biology Interface Training Grant (CBITG) Chemical Biology Colloquium Laboratory Rotation Program CBITG Areas of Training Bioorganic Chemistry Nucleic Acid Chemistry Med Chem Metallobiochemistry Chemistry Biocatalysis & Biomolecular Design BMBB Microbiology Engineering Metabolic Pathways Single Cell/ Molecule Studies Chemical Biology Coursework at U of M Flexible program Mechanisms of Chemical Reactions Introduction to Chemical Biology Chemical Biology of Enzymes Nucleic Acids Biochemistry Optional courses (2) Introduction to Graduate Research: Chem 8025 Course Objective Broaden exposure of students to new areas of chemistry by giving them practical hands-on exposure to graduate research in a participating faculty’s lab Course Description Rotate through two different laboratories. Each rotation lasts approximately seven weeks, with one credit earned for each rotation Research Areas Represented among Chemical Biology Faculty at U of M • Bioanalytical & Biomaterials • Computational Chemistry • Design & Synthesis • Enzyme Chemistry • Structure & Spectroscopy • Nucleic Acids Bioanalytical & Biomaterials Edgar Arriaga Michael Bowser Marian Stankovich Andrew Taton Subcellular distribution O 1 OH 11 12 10 9 2 3 8 5 4 6 OCH3 O H3C H OH 7 OH O 5' Doxorubicin 1' O 3' COCH2OH 4' OH NH2 2' 1.2 Metabolites Arriaga Fluorescence (V) 1.0 0.8 0.6 Group, 2005 http://www.chem.umn.edu/ groups/arriaga/ 0.4 0.2 100 150 200 Migration Time (s) 250 Bowser Group Research Interests http://www.chem.umn.edu/groups/bowser/ APTAMERS NEUROSCIENCE • • • • • • 10-Second In Vivo Monitoring Novel Neuromessengers Stroke Sensory Response New Sampling Methods Single Neuron Analysis • • • • Alternative Selection Methods (CE-SELEX) Microfluidics Catalytic Aptamers Aptamer Assays (neuropeptides, bacteria, etc.) Taton Group Research: Connecting Nanotech and Biotech http://www.chem.umn.edu/groups/taton/ Challenge: How to combine nanoobjects and biological molecules? Application Example: Nanoparticle labels for electrophoretic analysis of DNA (Electron microscope image of polymer-coated nanoparticles) (cuvette of DNA-modified, polymer-coated nanoparticles) DNA bands are visible with the naked eye. No staining necessary. Computational Chemistry Christopher Cramer (*) Jiali Gao William Gleason Donald Truhlar Rick Wagner Darrin York Jiali Gao http://www.chem.umn.edu/groups/gao Computational Biology • dynamics, pathways, and catalysis • protein-membrane interactions • macromolecular assembly Approach • combined QM/MM methods • MOVB • Monte Carlo and molecular dynamics Bill Gleason - Chemical Biology Interests Biomolecular recognition (e.g. heparin/protein interactions, receptor kinases) Biomaterials - synthesis and properties of novel polymeric biomaterials Biomarker discovery for clinical applications http://ccgb.umn.edu/~bgleason/ Mohammaddi Structure FGF:Receptor:Heparin 2:2:2 Truhlar Research Group CA H CD http://comp.chem.umn.edu/truhlar/ York Group: Density-functional theory and hybrid QM/MM methods Hybrid QM/MM simulations of reactions in RNA and in solution Density-functional calculations of biological reaction models http://riesling.chem.umn.edu/ Design & Synthesis George Barany Mark Distefano Craig Forsyth Larry Que Bill Tolman Rick Wagner Laundry Poker Proteins What do Laundry, Poker, and Proteins have in common? Laundry Poker Proteins A SYNTHETIC CHEMIST LOOKS AT PROTEIN FOLDING Prof. George Barany www.chem.umn.edu/ groups/baranygp/index.htm GB1 Distefano Research Group Organic and Protein Chemistry OH • Cancer N N 4' • Catalysis 5' • Antibiotics L38K S S V60C • Tissue Engineering http://www.chem.umn.edu/groups/distefano/ Forsyth Research Group PP1 - Okadaic Acid Docking Model V. A. Frydrychoski K. A. Plummer Wagner Research Group Interests Drug Design and Delivery Biocatalysis C-69 Chemical Biology -Antivirals -Antitumor D-122 H-107 -Carcinogen Activating Enzymes Nanobiotechnology -Chemical Control of Protein Macrocyclization Enzyme Chemistry Mark Distefano Craig Forsyth Jiali Gao John Lipscomb Karin Musier-Forsyth Larry Que Musier-Forsyth Lab I. Translation of the Genetic Code Species-specific differences in tRNA and amino acid recognition Expanding the genetic code:Incorporation of unnatural amino acids Amino acid editing/proofreading http://www.chem.umn.edu/groups/musier-forsyth/ Larry Que: Spectroscopic studies of oxygen activating iron enzymes with a 2-His-1-carboxylate motif • Enzymes catalyze hydrocarbon oxidations and antibiotic synthesis, degrade aromatic pollutants in soil, and sense hypoxia in cells • Spectroscopic methods used include EPR, NMR, Raman and EXAFS. Rapid kinetics techniques are used to trap reactive intermediates http://bioinorg.chem.umn.edu/quespace/ Structure & Spectroscopy John Lipscomb Karin Musier-Forsyth Larry Que Gianluigi Veglia QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. http://www.chem.umn.edu/groups/veglia/ NMR Studies of Membrane Proteins. Veglia Group Solution NMR Solid-state NMR Nucleic Acids Victor Bloomfield Michael Bowser Karin Musier-Forsyth Andrew Taton Darrin York Solid-Phase RNA/DNA Synthesizer Musier-Forsyth II. RNA-Protein Interactions in HIV Human tRNALys primer selection RNA chaperone activity of HIV nucleocapsid protein http://www.chem.umn.edu/groups/musier-forsyth/ York Group: Multi-scale Quantum Models for RNA catalysis External potential of solute and solvent Stochastic boundary Reaction Region QM active site + MM surrounding (Newtonian dynamics) Buffer Region (Langevin dynamics) http://riesling.chem.umn.edu/ Summary & Conclusions Chemical Biology: represents the “happening” interface for 21st century research Come to University of Minnesota, and help put the fun into “structure & function”! http://www.chem.umn.edu/bio/