* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Integrative Assignment - California State University
Expression vector wikipedia , lookup
Interactome wikipedia , lookup
Magnesium transporter wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
NADH:ubiquinone oxidoreductase (H+-translocating) wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Point mutation wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Biosynthesis wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Specialized pro-resolving mediators wikipedia , lookup
Homology modeling wikipedia , lookup
Genetic code wikipedia , lookup
Western blot wikipedia , lookup
Biochemistry wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Proteolysis wikipedia , lookup
Electron transport chain wikipedia , lookup
Metalloprotein wikipedia , lookup
Oxidative phosphorylation wikipedia , lookup
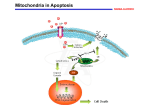
Integrative Assignment Part I Give a description of your organism. Homo sapiens is the primate species to which all modern humans belong. The closest relatives are chimpanzees from which they are distinguished by greater size, a peculiar distribution of hair, and a larger carbon footprint. Humans are primarily terrestrial animals and can be found on all continents. They are possibly the most thoroughly studied organism although mice, E. coli, and Drosophila are also in the running. (Kibak, Personal Communication). We study humans because we are self-obsessed. Domain: Eukaryota Kingdom: Metazoa Superphylum: Deuterostomia Phylum: Chordata Subphylum: Craniata Superclass: Euteleostomi Class: Mammalia Subclass: Eutheria Superorder: Euarchontoglires Order: Primates Suborder: Haplorrhini Superfamily: Catarrhini Family: Hominidae Genus: Homo Species: sapiens Figure above is from the National Geographic Genographic Project. Use an initial upper-case letter with the Genus name, initial lower-case letter with the species name. BOTH (and all foreign words) are italicized. Why organisms are studied: A. Ease of rearing or culture (e.g. Tribolium Red Flour Beetle, mouse, Arabidopsis, E. coli, Drosophila) B. Important in agriculture (Potato, Cow, Gibberella zeae) C. Important in medicine or disease (Penicillium marneffei ,Aedes aegypti, Ixodes scapularis) D. Important in industry (Kluyveromyces lactis, Yarrowia lipolytica) E. Extremist in some way (Emperor Penguin, Elephant Seal, Canis lupus familiaris) F. Ideal for one type of question for some reason (Sea Urchin, Neurospora, Danio rerio, Mytilus spp. ) Where in the cell is Cytochrome c found? What does Cytochrome c do? Cytochrome c diffuses back and forth between Complexes III and IV, being reduced by electrons from Complex III and oxidizing Complex IV as it offloads the electrons. See texbook, (Freeman, Biological Science). Cytochrome c is also a key protein in the regulation of apoptosis (Jiang & Wang, 2004) Cytochrome c (pink) Outer Mitochondrial Membrane Inter-membrane space Inner Mitochondrial Membrane Mitochondrial Matrix – Krebs Cycle Image courtesy of Research Collaboratory for Structural Bioinformatics “Molecule of the Month!” Cytochrome c is a peripheral membrane protein found on the inner mitochondrial membrane as a part of the Electron Transport Chain. Cytochrome c accepts electrons from Cytochrome c1 of Complex III and transfers them to Cytochrome a of Complex IV (also known as Cytochrome c oxidase). • Using the amino acid single letter code, write the primary structure of the Cytochrome b found in your organism. (citation) • How many amino acids are contained in Cytochrome b from your organism? To Answer Those Questions You Could: Google “cytochrome c” amino acid sequence Aedes aegypti to find information or use Google Scholar: But you will probably find too much advanced material to wade through… NCBI • Instead of using Google and the web, a better approach is to use the National Center for Biotechnology Information (NCBI) website. • NCBI is essentially “Google” for DNA and Protein sequences. Directions to Find the Cytochrome c for Your Organism • Google NCBI • In the NCBI’s front page search box type in species name and Cytochrome c • To the right, in the drop down menu, switch from “all databases” to “protein” and then click search. • Look through the search results starting at the top. You need to find a cytochrome C that is about 105aa long. It will say near the top of each article. Your protein WILL NOT BE “Cytochrome c Oxidase” – you will also have to retrieve that one for this assignment, but a bit later. • Once you find the right protein (about 100-120 aa’s long and correct species), switch GenPept to FASTA, you now have a more useable format. • Copy and paste to a notebook document. Save this file for for later. Copy and past the nicely formatted courier font sequence into your WORD document. This is a good one This one is too short FASTA is the format you want for your analysis. Note that all Cytochromes c have 95-120 amino acids. So here is the human Cytochrome c amino acid sequence… You need to do the same for your organism. This is the FASTA format for human Cytochrome c. It is displayed in a non-proportional font called “courier.” All your sequence data MUST BE DISPLAYED in COURIER. FASTA format has two requirements. 1. The first line has a “>” and contains information. 2. The following lines are sequence. >gi|11128019|ref|NP_061820.1| cytochrome c [Homo sapiens] MGDVEKGKKIFIMKCSQCHTVEKGGKHKTGPNLHGLFGRKTGQAPGYSYTAANKNKGIIWGE DTLMEYLENPKKYIPGTKMIFVGIKKKEERADLIAYLKKATNE How many amino acids are contained in Cytochrome c from your organism? What proportion of these amino acids are hydrophobic? Provide the best illustration you can for the tertiary structure of human Cytochrome c. Strategy 1. Take the human Cytochrome c sequence and search the structure (PDB) database in NCBI for similar sequences for which the structure has been solved. 2. Download Cn3D to display the structure. 3. Export the structure as a PNG file. 1. Select protein blast at NCBI BLAST http://www.ncbi.nlm.nih.gov/BLAST/ 2. Paste sequence. 3. Select “Protein Data Bank” 4. Select “blastp” Be PATIENT! The page will sit here for a while… updating several times until suddenly the results page appears. Click on the first structure link and see what it is… If it makes sense, then click on the S. First you will see this page. Then you will see this page If you don’t have Cn3D installed, download it. You won’t need admin privileges. It is a lot easier to work with peripheral membrane proteins than integral membrane proteins. Crystal structures have been solved for Cytochrome c from many organisms. http://www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi?db=t&form=6&dopt=s&uid=27574 If link doesn’t work, go to next page… Just like Cytochrome b, Cytochrome c has a heme group that is good at binding and releasing electrons. Where in the cell is Cytochrome c Oxidase found? What does Cytochrome c Oxidase do? Cytochrome c Oxidase is a protein in Complex Four of the Electron Transport Chain… would that make it an integral membrane protein? Human Cytochrome c Oxidase Human mitochondrion http://www.ncbi.nlm.nih.gov/nuccore/251831106?report=graph >ref|YP_003024028.1 cytochrome c oxidase subunit I Homo sapiens 513aa MFADRWLFSTNHKDIGTLYLLFGAWAGVLGTALSLLIRAELGQPGNLLGNDHIYNVIVTAHAFVMIFFMV MPIMIGGFGNWLVPLMIGAPDMAFPRMNNMSFWLLPPSLLLLLASAMVEAGAGTGWTVYPPLAGNYSHPG ASVDLTIFSLHLAGVSSILGAINFITTIINMKPPAMTQYQTPLFVWSVLITAVLLLLSLPVLAAGITMLL TDRNLNTTFFDPAGGGDPILYQHLFWFFGHPEVYILILPGFGMISHIVTYYSGKKEPFGYMGMVWAMMSI GFLGFIVWAHHMFTVGMDVDTRAYFTSATMIIAIPTGVKVFSWLATLHGSNMKWSAAVLWALGFIFLFTV GGLTGIVLANSSLDIVLHDTYYVVAHFHYVLSMGAVFAIMGGFIHWFPLFSGYTLDQTYAKIHFTIMFIG VNLTFFPQHFLGLSGMPRRYSDYPDAYTTWNILSSVGSFISLTAVMLMIFMIWEAFASKRKVLMVEEPSM NLEWLYGCPPPYHTFEEPVYMKS Source: http://www.rcsb.org/pdb/101/motm.do?momID=36 You can use wikipedia but you can’t cite it. Citation style = APA (see http://www.library.cornell.edu/resrch/citmanage/apa ) Freeman, S. (2011). Biological Science (4th Ed.). San Francisco: Pearson Benjamin Cummings. Jiang X, & Wang X. (2007). Cytochrome C-mediated apoptosis. Annual Review of Biochemistry, 73, 87-106. Mitochondrial ATP Synthase http://classes.csumb.edu/BIO/BIO241-01/campus/Lecture10respiration/ATPase.flv http://multimedia.mcb.harvard.edu/anim_ATPase_flv.html http://carbon.cudenver.edu/~bstith/1418m.mov