* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Document
Gene expression wikipedia , lookup
Signal transduction wikipedia , lookup
Magnesium transporter wikipedia , lookup
Biochemistry wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Interactome wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Metalloprotein wikipedia , lookup
Western blot wikipedia , lookup
Structural alignment wikipedia , lookup
Homology modeling wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Protein–protein interaction wikipedia , lookup
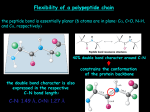
Principles of Protein Structure primary structure ACDEFGHIKLMNPQRSTVWY Different Levels of Protein Structure NH2 Lysine Histidine Valine Arginine Alanine COOH Common Secondary Structure Elements • The Alpha Helix Properties of alpha helix • • • • • • • 3.6 residues per turn, 13 atoms between H-bond donor and acceptor approx. -60º; approx. -40º H- bond between C=O of ith residue & -NH of (i+4)th residue First -NH and last C=O groups at the ends of helices do not participate in Hbond Ends of helices are polar, and almost always at surfaces of proteins Always right- handed Macro- dipole Alpha Helix Helical wheel Residues i, i+4, i+7 occur on one face of helices, and hence show definite pattern of hydrophobicity/ hydrophilicity Association of helices: coiled coils Introduction to Molecular Biophysics These coiled coils have a heptad repeat abcdefg with nonpolar residues at position a and d and an electrostatic interaction between residues e and g. Isolated alpha helices are unstable in solution but are very stable in coiled coil structures because of the interactions between them The chains in a coiled-coil have the polypeptide chains aligned parallel and in exact axial register. This maximizes coil formation between chains. The coiled coil is a protein motif that is often used to control oligomerization. They involve a number of alpha-helices wound around each other in a highly organised manner, similar to the strands of a rope. The Leucine Zipper Coiled Coil Introduction to Molecular Biophysics Initially identified as a structural motif in proteins involved in eukaryotic transcription. (Landschultz et al., Science 240: 1759-1763 (1988). Originally identified in the liver transcription factor C/EBP which has a Leu at every seventh position in a 28 residue segment. Association of helices: coiled coils The helices do not have to run in the same direction for this type of interaction to occur, although parallel conformation is more common. Antiparallel conformation is very rare in trimers and unknown in pentamers, but more common in intramolecular dimers, where the two helices are often connected by a short loop. Chan et al., Cell 89, Pages 263-273. Basis for the helical dipole In an alpha helix all of the peptide dipoles are oriented along the same direction. Consequently, the alpha helix has a net dipole moment. Since the dipole moment of a peptide bond is 3.5 Debye units, the alpha helix has a net macrodipole of: n X 3.5 Debye units (where n= number of residues) This is equivalent to 0.5 – 0.7 unit charge at the end of the helix. The amino terminus of an alpha helix is positive and the carboxy terminus is negative. Structure of human TIM Two helix dipoles are seen to play important roles: 1. 2. Stabilization of inhibitor 2-PG Modulation of pKa of active site His-95. Helical Propensities Ala Arg Lys Leu Met Trp Phe Ser Gln Glu Cys Ile Tyr Asp Val Thr Asn His Gly Pro -0.77 -0.68 -0.65 -0.62 -0.50 -0.45 -0.41 -0.35 -0.33 -0.27 -0.23 -0.23 -0.17 -0.15 -0.14 -0.11 -0.07 -0.06 0 ~3 Common Secondary Structure Elements • The Beta Sheet Secondary structure: reverse turns Secondary Structure: Phi & Psi Angles Defined • Rotational constraints emerge from interactions with bulky groups (ie. side chains). • Phi & Psi angles define the secondary structure adopted by a protein. The dihedral angles at C atom of every residue provide polypeptides requisite conformational diversity, whereby the polypeptide chain can fold into a globular shape Ramachandran Plot Secondary Structure Table 10 Phi & Psi angles for Regular Secondary Structure Conformations Structure Antiparallel b-sheet Parallel b-Sheet Right-handed -helix 310 helix p helix Polyproline I Polyproline II Polyglycine II Phi (F) -139 -119 +64 -49 -57 -83 -78 -80 Psi(Y) +135 +113 +40 -26 -70 +158 +149 +150 Beyond Secondary Structure Supersecondary structure (motifs): small, discrete, commonly observed aggregates of secondary structures b sheet helix-loop-helix bb Domains: independent units of structure b barrel four-helix bundle *Domains and motifs sometimes interchanged* Common motifs Supersecondary structure: Crossovers in b--b-motifs Left handed Right handed EF Hand • Consists of two perpendicular 10 to 12 residue alpha helices with a 12-residue loop region between • Form a single calcium-binding site (helix-loop-helix). • Calcium ions interact with residues contained within the loop region. • Each of the 12 residues in the loop region is important for calcium coordination. • In most EF-hand proteins the residue at position 12 is a glutamate. The glutamate contributes both its side-chain oxygens for calcium coordination. Calmodulin, recoverin : Regulatory proteins Calbindin, parvalbumin: Structural proteins EF Fold Found in Calcium binding proteins such as Calmodulin Helix Turn Helix Motif •Consists of two helices and a short extended amino acid chain between them. •Carboxyl-terminal helix fits into the major groove of DNA. •This motif is found in DNA-binding proteins, including l repressor, tryptophan repressor, catabolite activator protein (CAP) Leucine Zipper Rossman Fold •The beta-alpha-beta-alpha-beta subunit •Often present in nucleotide-binding proteins What is a Protein Fold? Compact, globular folding arrangement of the polypeptide chain Chain folds to optimise packing of the hydrophobic residues in the interior core of the protein Common folds Tertiary structure examples: All- Cytochrome C four-helix bundle Alamethicin The lone helix Rop helix-turn-helix Tertiary structure examples: All-b b sandwich b barrel Tertiary structure examples: /b placental ribonuclease inhibitor /b horseshoe triose phosphate isomerase /b barrel Four helix bundle •24 amino acid peptide with a hydrophobic surface •Assembles into 4 helix bundle through hydrophobic regions •Maintains solubility of membrane proteins Oligonucleotide Binding (OB) fold TIM Barrel •The eight-stranded /b barrel (TIM barrel) •The most common tertiary fold observed in high resolution protein crystal structures •10% of all known enzymes have this domain Zinc Finger Motif Domains are independently folding structural units. Often, but not necessarily, they are contiguous on the peptide chain. Often domain boundaries are also intron boundaries. Domain swapping: Parts of a peptide chain can reach into neighboring structural elements: helices/strands in other domains or whole domains in other subunits. Domain swapped diphteria toxin: Transmembrane Motifs • Helix bundles Long stretches of apolar amino acids Fold into transmembrane alpha-helices “Positive-inside rule” Cell surface receptors Ion channels Active and passive transporters • Beta-barrel Anti-parallel sheets rolled into cylinder Outer membrane of Gram-negative bacteria Porins (passive, selective diffusion) Quaternary Structure • Refers to the organization of subunits in a protein with multiple subunits • Subunits may be identical or different • Subunits have a defined stoichiometry and arrangement • Subunits held together by weak, noncovalent interactions (hydrophobic, electrostatic) • Associate to form dimers, trimers, tetramers etc. (oligomer) • Typical Kd for two subunits: 10-8 to 10-16M (tight association) –Entropy loss due to association - unfavorable –Entropy gain due to burying of hydrophobic groups - very favourable Structural and functional advantages of quaternary structure • • • • Stability: reduction of surface to volume ratio Genetic economy and efficiency Bringing catalytic sites together Cooperativity (allostery) Quaternary structure of multidomain proteins