* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download THERAPUETIC DISCOVERY BY MODELLING
Structural alignment wikipedia , lookup
Rosetta@home wikipedia , lookup
List of types of proteins wikipedia , lookup
Protein folding wikipedia , lookup
Protein domain wikipedia , lookup
Bimolecular fluorescence complementation wikipedia , lookup
Homology modeling wikipedia , lookup
Western blot wikipedia , lookup
Degradomics wikipedia , lookup
Protein purification wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Protein structure prediction wikipedia , lookup
Protein mass spectrometry wikipedia , lookup
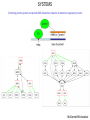
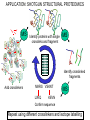
THERAPUETIC DISCOVERY BY MODELLING INTERACTOMES RAM SAMUDRALA ASSOCIATE PROFESSOR UNIVERSITY OF WASHINGTON How does the genome of an organism specify its behaviour and characteristics? How can we capitalise on this information to discover therapeutics to improve human health and quality of life? STRUCTURE T0290 – peptidyl-prolyl isomerase from H. sapiens T0288 – PRKCA-binding from H. sapiens 0.5 Å Cα RMSD for 173 residues (60% identity) 2.2 Å Cα RMSD for 93 residues (25% identity) T0332 – methyltransferase from H. sapiens T0364 – hypothetical from P. putida 2.0 Å Cα RMSD for 159 residues (23% identity) 5.3 Å Cα RMSD for 153 residues (11% identity) Liu/Hong-Hung/Ngan FUNCTION Ion binding energy prediction with a correlation of 0.7 Calcium ions predicted to < 0.05 Å RMSD in 130 cases Meta-functional signature accuracy Meta-functional signature for DXS model from M. tuberculosis Wang/Cheng INTERACTION Transcription factor bound to DNA promoter regulog model from S. cerevisiae Prediction of binding energies of HIV protease mutants and inhibitors using docking with dynamics BtubA/BtubB interolog model from P. dejongeii (35% identity to eukaryotic tubulins) McDermott/Wichadakul/Staley/Horst/Manocheewa/Jenwitheesuk/Bernard SYSTEMS Example predicted protein interaction network from M. tuberculosis (107 proteins with 762 unique interactions) Proteins PPIs TRIs H. sapiens 26,741 17,652 828,807 1,045,622 S. cerevisiae 5,801 5,175 192,505 2,456 O.sativa (6) 125,568 19,810 338,783 439,990 E. coli 4,208 885 1,980 54,619 In sum, we can predict functions for more than 50% of a proteome, approximately ten million protein-protein and protein-DNA interactions with an expected accuracy of 50%. Utility in identifying function, essential proteins, and host pathogen interactions McDermott/Wichadakul SYSTEMS Combining protein-protein and protein-DNA interaction networks to determine regulatory circuits McDermott/Wichadakul INFRASTRUCTURE ~500,000 molecules over 50+proteomes served using a 1.2 TB PostgreSQL database and a sophisticated AJAX webapplication and XML-RPC API http://bioverse.compbio.washington.edu http://protinfo.compbio.washington.edu Guerquin/Frazier INFRASTRUCTURE Guerquin/Frazier INFRASTRUCTURE http://bioverse.compbio.washington.edu/integrator Chang/Rashid APPLICATION: DRUG DISCOVERY Drug discovery as undertaken by the pharmaceutical company is time consuming and expensive, with very low hit rates for the amount of resources expended. Computational screening of compounds against structures of protein targets offers a way to speed up discovery time and reduce costs, but such techniques have typically had low accuracy and need high resolution structures. We will capitalise on advances in computational protein structure prediction and protein docking to improve accuracy of target-based in silico compound screening. APPLICATION: DRUG DISCOVERY HSV CMV KHSV Jenwitheesuk APPLICATION: DRUG DISCOVERY Computionally predicted broad spectrum human herpesvirus protease inhibitors is effective in vitro against members from all three classes and is comparable or better than anti-herpes drugs HSV KHSV CMV Our protease inhibitor acts synergistically with acylovir (a nucleoside analogue that inhibits replication) and it is less likely to lead to resistant strains compared to acylovir HSV HSV Lagunoff Multitarget inhibition of Plasmodium falciparum proteins Ekachai Jenwitheesuk/Wesley Van Voorhis Multi-target inhibition of Plasmodium falciparum proteins We experimentally evaluated 16 of our top predictions against P. falciparum in cell culture. 6/16 had an ED50 of 1 M, with the best inhibitor having an ED50 of 127nM. A negative control of 5 randomly selected compounds predicted to not inhibit our fourteen targets did not inhibit P. falciparum growth. Chong et al.1 experimentally screened 2687 compounds and found 87 inhibitors against P. falciparum. Weisman et al.2 screened 2162 compounds found 72 inhibitors. Their hit rates are 3.2% (87/2687) and 3.3% (72/2162). We are thus able to obtain a much higher hit rate of 38% (6/16) for a fraction of the cost: Only 16 compounds costing ~$1000 needed to be tested. Computation is fully automated and takes only a few days. Examining overlap between our computational library and their experimental libraries resulted in 75 compounds of which we would have tested 15. 8/15 inhibitors had an ED50 of 1M, resulting in a hit rate of 53%. 1Nat Chem Biol 2: 415-6, 2006. Biol Drug Des 409-16, 2006. 2Chem Ekachai Jenwitheesuk/Wesley Van Voorhis Other work and future directions Our predicted inhibitors against the dengue virus are more efficacious in cell culture than previously identified inhibitors We have predicted inhibitors against more than 100 protein targets for over 20 diseases, including HIV, SARS, Leishmania, Tuberculosis, and Influenza. Experimental testing is underway against some of the pathogens responsible. Computationally screen structurally-related compounds to experimentally verified inhibitors from a much larger library of 1 million compounds. Use data from experimental studies to figure out when our predicted inhibitors are likely to be cell-active and drug-like in their behaviour; use machine learning approaches to learn from compound characteristics (PK, ADME, toxicity), importance of protein targets, predicted binding energies and experimental inhibition. Works due to the use of a combination of knowledge- and biophysics-based methods for computational simulation. APPLICATION: NANOTECHNOLOGY Oren/Sarikaya/Tamerler APPLICATION: SHOTGUN STRUCTURAL PROTEOMICS MS Identify proteins with single crosslinks and fragment MS Identify crosslinked fragments Add crosslinkers MKRS VSKNT MS LVKQ KEVN Confirm sequence Repeat using different crosslinkers and isotope labelling ACKNOWLEDGEMENTS Current group members: Past group members: •Baishali Chanda •Brady Bernard •Chuck Mader •David Nickle •Ersin Emre Oren •Ekachai Jenwitheesuk •Gong Cheng •Imran Rashid •Jeremy Horst •Ling-Hong Hung •Michal Guerquin •Rob Brasier •Rosalia Tungaraza •Shing-Chung Ngan •Siriphan Manocheewa •Somsak Phattarasukol •Stewart Moughon •Tianyun Liu •Vania Wang •Weerayuth Kittichotirat •Zach Frazier •Kristina Montgomery, Program Manager •Aaron Chang •Duncan Milburn •Jason McDermott •Kai Wang •Marissa LaMadrid Collaborators: •James Staley •Mehmet Sarikaya/Candan Tamerler •Michael Lagunoff •Roger Bumgarner •Wesley Van Voorhis Funding agencies: •National Institutes of Health •National Science Foundation •Searle Scholars Program •Puget Sound Partners in Global Health •UW Advanced Technology Initiative •Washington Research Foundation •UW TGIF