* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Course Outline
Heritability of autism wikipedia , lookup
Gene expression programming wikipedia , lookup
Genetic engineering wikipedia , lookup
Hardy–Weinberg principle wikipedia , lookup
History of genetic engineering wikipedia , lookup
Genetic testing wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Genetics and archaeogenetics of South Asia wikipedia , lookup
Dual inheritance theory wikipedia , lookup
Pharmacogenomics wikipedia , lookup
Koinophilia wikipedia , lookup
Designer baby wikipedia , lookup
Medical genetics wikipedia , lookup
Dominance (genetics) wikipedia , lookup
Public health genomics wikipedia , lookup
Genome (book) wikipedia , lookup
Genetic drift wikipedia , lookup
Polymorphism (biology) wikipedia , lookup
Human genetic variation wikipedia , lookup
Behavioural genetics wikipedia , lookup
Microevolution wikipedia , lookup
Population genetics wikipedia , lookup
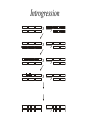
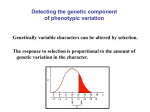
Course Outline 1. 2. 3. 4. 5. 6. Introduction: QTL and heritability Quantitative Mutagenesis QTL Mapping QTN Mapping Microarrays and Genomics Evolutionary genetics Introduction • Genetic analysis of common human diseases, animal and plant breeding, and evolutionary biology all require the tools of quantitative genetics. • Quantitative genetics is the study of the variation in continuous traits. • Small effects of many individual genes add together and interact with the environment, to produce natural variation • Modern molecular quantitative genetics is focused on identifying the underlying genes and describing how variation at the DNA level translates into phenotypic variation Oligogenic effects 1 Locus Frequency Frequency 2 Loci 1 2 - + - 1 + + 1 4 - - - + - - Frequency 3 Loci 1 - - - - - 6 15 20 15 + + - - - - - + + + - - + - - + - - + - + + - + - - + + + + - - 6 1 + + + + + - - + + + + + + + - + + + - 6 + + + + - 4 + + + - 1 + + + + Heritability • Heritability is the proportion of phenotypic variation in a population that is due to genotypic variation • Heritability describes the relative contributions of genotype and environment to phenotypic variation • Heritability is a measure of a population, not an individual, and does not say anything about the genetic difference between populations. • There are three common approaches to measuring heritability. Parent-Offspring Regression Offspring Phenotype 6 5 4 3 2 1 1 2 3 4 Parental Phenotype 5 6 Variance among Lines High heritability 6 6 5 5 4 4 Phenotype Phenotype Low heritability 3 3 2 2 1 1 1 2 3 Line Number 4 1 2 3 Line Number 4 Realized heritability Frequency Initial population Selected sample O R S R Progeny of selected sample Phenotype S Polygenes • The number of genes that affect a trait can be estimated from the variance segregating in an F2 generation • There is a wide range in the number of genes that affect traits • As the number of genes influencing a trait increases, the relative number of individuals with extreme phenotypes decreases. • Transgressive segregation occurs when grandchildren have more extreme phenotypes than either grandparent. Simple additive model ¼ AA (0) ½ Aa (1) ¼ aa (2) ¼ BB (0) 1/16 (0) 1/8 (1) 1/16 (2) ½ Bb (1) 1/8 (1) 1/4 (2) 1/8 (3) ¼ Bb (2) 1/16 (2) 1/8 (3) 1/16 (4) Polygenic effects Frequency Parental samples Frequency F2 samples Frequency F1 sample Phenotype Transgressive segregation Parental samples Frequency A. - ; + + ; -- ; - + + ; - ; ++ ; + + B. F2 F1 F2 samples Frequency P2 Frequency P1 25 35 45 55 65 75 85 95 105 115 Plant height (cm) - ; - ; -- ; - + + Phenotype ; + + ; ++ ; + + VP = VA + VD + VI + VGxE + VE • Loci are said to have Additive effects if the contributions of each individual allele can simply be added algebraically to arrive at a prediction of a phenotype given a genotype. • Dominance refers to the observation that heterozygotes resemble one class of homozygotes more than the other. • Epistasis refers to a locus-by-locus Interaction, such as when alleles at two loci antagonize or synergize with one another. • VE is the environmental variance Mean phenotype Dominance ratio 20 18 16 14 12 10 8 6 4 2 0 bb Bb a d d = 16-10 = 6 a = 18-10 = 8 0 Expected mid-value = (18+2)/2 = 10 BB -a 0 1 Number of “b” alleles 2 Epistasis OBSERVED 38 37 36 35 34 33 32 31 EXPECTED 38 37 36 35 34 33 32 31 BB Bb bb AA Aa BB Bb bb aa AA EPISTATIC DEVIATIONS 1.00 0.75 0.50 0.25 0.00 - 0.25 - 0.50 - 0.75 - 1.00 Bb bb BB AA Aa aa Aa aa Quantitative Mutagenesis • Quantitative genetic variation is constantly produced by mutation accumulation • Quantitative measurement of P-elements can uncover novel genes regulating complex traits • Deficiency complementation mapping is a way to infer that segregating variation affects a trait Mutation accumulation H1 H2 H3 Number of Bristles 20 15 10 5 0 20 L1 L2 L3 40 60 80 Generation 100 120 P screen for bristle number Df Complementation Df wt X Vs wt wt Bal wt 140.00 120.00 Survival Time Df Bal Females 100.00 80.00 60.00 40.00 20.00 0.00 Mutant Wildtype Introgression X X X X omb bs/DSRF Gap1 sbb ptc ed egfr star osa drk Argos spi dad mam Mad ksr pnt tkv Trl rho- 2 dpp ast src42A sd mam cbl tkv cos mad psq cv-2 dpp babo drk brinker sax tsh sax PKA-C-3 RAS85D CG3957 rho1 kinsahn se rho- AP PKA p38b rho- 6 cs RAS-GAw P rho- 6 -LOG(P) P’s affect wing shape 50 45 40 GENOTYPE GENOTYPE*BACKGROUND 35 30 25 20 15 10 5 0 LINE QTL Mapping • A QTL is a locus that has a quantitative effect on a trait • QTL mapping is similar to linkage mapping for Mendelian traits, except that genotypes are only correlated with phenotypes • In the laboratory, F2, Backcross and RIL designs are commonly employed to map QTL • Molecular markers include RFLPs, Microsatellites, and SNPs Principle of QTL mapping + + + + - + - - - - + + X - + - 112 cm - + - - + + - - - + + + + - 94 cm - 88 cm - + + - + - - - 100 cm - + + - + - 93 cm - 101 cm A QTL Profile QTL Mapping Designs A. F2 design X X B. BC design X C. RIL design X X Types of Molecular Marker A. Single nucleotide polymorphism ACCGTTCAGCAGATCAG TGGCAAGTCGTCTAGTC ACCGTTCAACAGATCAG TGGCAAGTTGTCTAGTC B. Indel Polymorphism ACCGTTCAGCAGATCAG TGGCAAGTCGTCTAGTC ACCGTTCA---GATCAG TGGCAAGT---CTAGTC C. Microsatellite polymorphism ACCGTGTGTGTGTTCAG TGGCACACACACAAGTC ACCGTGTGTGTGTGTTCAG TGGCACACACACACAAGTC Likelihood Ratio Likelihood Ratio Interval Mapping 0 20 40 60 80 Position (cM) 100 0 20 40 60 80 Position (cM) 100 QTN Mapping • Linkage disequilibrium (LD) mapping uses all of the recombinational history in a population to increase the resolution of QTL mapping • Modern human quantitative genetics utilizes the HapMap project for Association Mapping • Different organisms have different LD structure, so the power of QTL mapping is somewhat species-specific • In flies, there is so little LD that genome scans are precluded; however, resolution may be to the QTN (Quanitative Trait Nucleotide) level Principle of LD Mapping 1,000 generations Genotypes and Haplotypes Random Polymorphisms Haplotype Blocks 1 2 A G C A T A T C A G C C A C A G C A C C A A C T 1 2 A C C G T T C G A G C T A C C G C A T C A G C T 3 4 T C A G C A C G A G C T A C C A C T T G T G A T 3 4 T G A A C A T C A G C T A C C G C A T C T A A C 5 6 A G A G T A T C A G C T A G C A C A T C A A C C 2 4 A C C G C A T C A G C T A C C G C A T C T A A C 7 8 A C C G C A T C A G A T T G C G T T T C A G C T 2 5 A C C G C A T C A G C T T G A A T T C G A G C T 9 3 T C C A C T T C T G C T T C A G C A C G A G C T 3 5 T G A A C A T C A G C T T G A A T T C G A G C T LD in EGFR A QTN Profile 5 C1 4.5 30200 4 Significance 5 3.5 3.5 3 2.5 2.5 2 2 1.5 1.5 1 1 0.5 0.5 0 0 W1 5 4.5 Significance 39389 4 3 5 D1 4.5 40110 6065 W9 4.5 4 4 3.5 3.5 3 3 2.5 2.5 2 2 1.5 1.5 1 1 0.5 0.5 0 0 Location of SNP Location of SNP Quantitative Genomics • A collaborative cross has been proposed for fine-structure mapping of a wide range of traits in D. melanogaster and D. simulans • Microarrays have many applications from modeling the evolution of transcription to identifying candidate genes for quantitative traits • The future lies in network analysis Gene Expression Among Lines Adaptation • The relative roles of genetic drift, selection, mutation, migration and other evolutionary processes in biological evolution continue to be debated • Adaptive walks occur by the successive fixation of alleles, at least one of which is expected to have a major effect on the trait • Soft selection refers to adaptation that starts with hidden genetic variation whose effect on fitness appears when the environment changes, whereas hard selection starts from newly arising mutations Wright’s Shifting Balance Phase 1 Phase 2 Phase 3 An Adaptive Walk A. B. C. Canalization • Canalization is the evolution of buffering of physiological or developmental systems. • Homeostasis is buffering due to the structure of genetic networks, and/or the function of biochemical mechanisms • Canalization allows the build up of cryptic genetic variation that is available to modify phenotypes upon genetic or environmental perturbation Canalization Perturbed population Phenotype Frequency Canalized population Fundamental Questions • What maintains genetic variation in natural populations? • Is most quantitative genetic variation due to common alleles of small effect, or rare alleles of large effect? • What is the origin of dominance? • How pervasive is epistasis, and does it contribute meaningfully to evolution?