* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Mammal evolution and biogeography
Promoter (genetics) wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Ridge (biology) wikipedia , lookup
Gene regulatory network wikipedia , lookup
Genome evolution wikipedia , lookup
Community fingerprinting wikipedia , lookup
Genomic imprinting wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
Gene expression profiling wikipedia , lookup
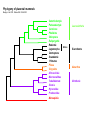
Mammalian Evolution Using IRBP Gene Goal: To provide a problem space wherein students can use sequence data using a slowly evolving genes to resolve deep-level divergences within and among mammalian groups. To illustrate the explanatory powers of (molecular) phylogenetics for biology. Problems with deep-level divergences Mitochondrial markers are too fast evolving to resolve divergences of more than >50 millions years. - Data sets often beset by saturation (homoplasy) and trees often suffer from long branch attractions. Some Potential Questions: Does the major grouping of mammals reflect biogeographic history? Does molecular phylogeny reflect present (worldwide or local) distribution? Are marsupials monophyletic relative to eutherian mammals? Are rodents monophyletic? To what groups of mammals whales and dolphins are most related to? Opportunities for students Provides students to pose many other questions and generate many hypotheses. Investigate the concept of HOMOLOGY in both molecular and morphological (or behavioral, ecological ) data sets Test geographic events with character evolution. Allow students to view biodiversity (and conservation) in the context of evolution. IRBP exon 1(Interphotoreceptor Binding Protein) • Single copy nuclear gene (currently the most widely used marker for mammalian phylogeny) • Widely used to infer mammalian phylogeny • Relative rate of mutation is slower than mitochondrial genes, thus, ideal marker to resolve deep-level divergences Phylogeny of placental mammals Murphy et al. 2001. Nature.294: 2348-2351 Cetartiodactyla Perissodactyla Carnivora Pholidota Chiroptera Eulipotyphla Rodentia Glires Lagomorpha Dermoptera Scandentia Primates Pilosa Cingulata Afrosoricida Macroscelidea Tubulidentata Sirenia Hyracoidea Proboscidea Marsupialia Laurasiatheria Euarchonta Xenarthra Afrotheria Other Possibilities Using other genes, such as vonWillebrand and rag-1 genes to further illustrate the virtues of the scientific concept: Testability, Repeatability, Congruence among independent data sets