* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Supplemental Information
Paracrine signalling wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Evolution of metal ions in biological systems wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Multi-state modeling of biomolecules wikipedia , lookup
Size-exclusion chromatography wikipedia , lookup
Cryobiology wikipedia , lookup
Signal transduction wikipedia , lookup
Biochemical cascade wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Western blot wikipedia , lookup
Polyclonal B cell response wikipedia , lookup
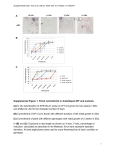
Supporting Information Novel anticancer agents based on targeting the trimer interface of the PRL phosphatase Yunpeng Bai1, Zhi-Hong Yu1, Sijiu Liu2, Lujuan Zhang2, Ruo-Yu Zhang1, Li-Fan Zeng2, Sheng Zhang1, and Zhong-Yin Zhang1,2,* 1 Department of Medicinal Chemistry and Molecular Pharmacology, Department of Chemistry, Purdue Center for Cancer Research, and Purdue Center for Drug Discovery, Purdue University, 575 Stadium Mall Drive, West Lafayette, IN 47907, USA; 2 Department of Biochemistry and Molecular Biology, Indiana University School of Medicine, 635 Barnhill Drive, Indianapolis, IN 46202, USA *To whom correspondence should be addressed: [email protected]. Running title: PRL detrimerizers as novel anticancer agents In vitro and in vivo cross-linking experiments. To assess the oligomeric state of PRL1 in vitro, (His)6-tagged recombinant PRL1 proteins were cross-linked with glutaraldehyde (Sigma, 25% glutaraldehyde solution, grade I). The reactions were performed in 20 μL solutions containing 5 μg of protein in phosphate-buffered saline (PBS, pH 7.5). Recombinant PRL1 was treated with 10 μM compound for 30 min, and then cross-linked by incubating with 0.005% glutaraldehyde at room temperature for 10 min. The reaction was terminated by addition of pH 7.5 Tris-HCl (final concentration 50 mM) and 5 min incubation on ice. The samples were separated on SDS-PAGE and analyzed by Coomassie Blue staining. For in vivo cross-linking, HEK293 cells with 90% confluence were treated with 20 μM compound for 24 hours, and then fixed with 1% formaldehyde (Thermo, 16% formaldehyde solution) for 10 min at room temperature. Cells were washed twice in ice-cold PBS and lysed on ice for 30 min following by immunoprecipitation with HA antibody. The reaction mixtures were analyzed by SDSPAGE and detected by immunoblotting with anti-HA antibody (Santa Cruz). The detrimerization efficacy of the compounds was determined by percentage of trimer/monomer ratio reduction. Synthesis of Cmpd-43 and its analogs. To synthesize methyl 3-(5,6dimethylisoindolin-2-yl)benzoate, a solution of 3-(5,6-dimethylisoindolin-2-yl)benzoic acid (267 mg, 1 mmol), concentrated H2SO4 (0.5 mmol) in methanol (20 mL) were heated to reflux overnight. The methanol was removed under vacuum. The mixture was quenched with saturated NaHCO3 (100 mL) and extracted with EtOAc (3 * 50 mL). The combined organic extract was dried over sodium sulfate and the solvent was removed in vacuum. The residue was purified by silica gel column chromatography using hexane/ethyl acetate (8:1) to provide the title compound as a pale solid (60 mg, 21.3%). To synthesize Analog-1 (3-(5,6-dimethylisoindolin-2-yl)benzohydrazide), a solution of methyl 3-(5,6-dimethylisoindolin-2-yl)benzoate (1.4 g, 4.98 mmol), hydrazine hydrate (3 mL) in methanol (20 mL) were heated to reflux for 3 h. Most of the methanol was removed under vacuum. The residue was washed by water (5 mL) and methanol (5 mL) to afford title compound as white solid (1.1 g, 78.5%). To synthesize Cmpd-43 and Analog-5 to Analog-7, to a stirred solution of Analog-1 (20 mg, 0.07 mmol) and appropriate benzaldehyde (0.15 mmol) in methanol (5 mL), glacial acetic acid (0.1 mL) was added. The mixture was refluxed for 3 h, after which the solution was cool down, filtrated and the resulting solid was washed by methanol (2 mL) to afford title compounds as pale solids. Assessment of PTP inhibition by Cmpd-43. PTP activity was assayed using pNPP as a substrate at 25C in 50 mM 3,3-dimethylglutarate buffer, pH 7.0, containing 1 mM EDTA with an ionic strength of 0.15 M adjusted by NaCl. The assays were performed in 96-well plates in a total reaction volume of 200 μL. The reaction was initiated by the addition of enzyme to a reaction mixture containing pNPP at a concentration equaling the Km value for each PTP and 20 M Cmpd-43. The reaction was quenched by the addition of 5 M NaOH. The nonenzymatic hydrolysis of pNPP was corrected by measuring the control without the addition of enzyme. The amount of product p-nitrophenol was determined from the absorbance at 405 nm detected by a Spectra MAX340 microplate spectrophotometer (Molecular Devices) using a molar extinction coefficient of 18,000 M1 cm-1. In addition to PRL1, other PTPs evaluated included the receptor-like PTPs, PTPμ, PTPε, LAR, PTPσ and PTPγ, cytosolic PTPs, PTP1B, Lyp, SHP1, PTPH1, HePTP, STEP, and PEZ, the dual specificity phosphatase VHR, VHZ, MKP5, CDC14A, and the low molecular weight PTP. PRL1 expression, purification, crystallization and data collection. Recombinant PRL1 expression and purification was described previously (1). For co-crystallization of PRL1 with Analog-3, 100 μL 6 mg/mL PRL1 was mixed with 6 μL stock of Analog-3 (10 mM in DMSO), then crystals were grown by vapor diffusion in hanging drops at 4C, room temperature, 30C and 37 C respectively. Drops containing 1:1 volumes of protein in stock buffer and reservoir solutions were equilibrated against the reservoir solution A (1.9 M ammonium sulfate, 100 mM sodium acetate, pH 4.6 ~ 4.9). The crystals were observed after 2 weeks (for 37 C), or 7 weeks (for 30 C), or half year (for room temperature); No crystals were observed under 4 C condition in 8 months. The crystals (grown at 37 C) was transferred into a reservoir solution B (90% saturated lithium sulfate, 0.5 mM Analog-3, 5% DMSO), soaked for 1 second, then flash-cooled in liquid nitrogen. X-ray data were collected at 100 K at SBC-CAT beamline 19-BM at the Advanced Photon Source (Argonne, IL) equipped with a mosaic CCD detector. The crystals belong to space group C2221 with the following unit cell parameters: a = 46.07 Å, b = 76.47 Å and c = 86.87 Å. There is one protein molecule in the asymmetric unit. All data were processed with HKL3000 (2), and the statistics are provided in Table 1. Structural determination and refinement. The structure of PRL1Analog-3 was solved by molecular replacement using the program Molrep (3) . The structure of PRL1 (PDB entry code 1ZCK) (4), without the solvent molecules and other small molecules, was used as a search model. The map revealed the density for the bound Analog-3 (Figure 4). The structure was refined to 1.90 Å resolution with the program CNS1.1 (5), first using simulated annealing at 2,500 K, and then alternating positional and individual temperature factor refinement cycles. Electron density maps were inspected and the model was modified in WinCoot (6). Finally, water molecules were added gradually as the refinement progressed. They were assigned in the |Fo| - |Fc| difference Fourier maps with a 3σ cutoff level for inclusion in the model. The statistics of refinements were also provided in Table 1. Molecular graphics were prepared using Pymol (www.pymol.sourceforge.net). Histological studies. Tumors and tissues were fixed in 4% paraformaldehyde (PFA) overnight at 4 °C, embedded in paraffin, serially sectioned (7 μm), and stained with H&E according to standard methods. For immunohistochemistry, de-paraffined and hydrated sections were subjected to antigen retrieval by boiling in 10 mM sodium citrate for 20 min. Sections were then incubated with diluted antibodies (1:50-1:400) at 4 °C overnight. Signals were detected by VECTASTAIN Elite ABC kit and developed using DAB substrate from Vector laboratory (Burlingame, CA). Antibodies used were Ki67 (Thermo Fisher Scientific, MA) and cleaved PARP (Cell Signaling Technology, MA). Images were captured on a Leica DM2500 stereomicroscope. All images are representative of at least 3 samples. References 1. Bai Y, Luo Y, Liu S, Zhang L, Shen K, Dong Y, et al. PRL-1 protein promotes ERK1/2 and RhoA protein activation through a non-canonical interaction with the Src homology 3 domain of p115 Rho GTPase-activating protein. J Biol Chem 2011;286:42316–24. 2. Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr 2006;62:859–66. 3. Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. Journal of Applied Crystallography 1997;30:1022–5 4. Sun JP, Wang WQ, Yang H, Liu S, Liang F, Fedorov AA, et al. Structure and biochemical properties of PRL-1, a phosphatase implicated in cell growth, differentiation, and tumor invasion. Biochemistry 2005;44:12009–21. 5. Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallographica Section D-Biological Crystallography 1998;54:905–21. 6. Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 2010;66:486–501. Supplemental Figure 1 Supplemental Figure 1. Cmpd-43 selectively inhibits wild-type MEF cells against PRL1-/MEF cells. *p<0.05 (Student's t test), data represent mean (SD) value from 3 independent experiments. Supplemental Figure 2 Supplemental Figure 2. Cmpd-43 showed less cell toxicity towards non-tumor cell lines. A. Compared to breast cancer cell line MCF7, the cell toxicity of Cmpd-43 was significantly attenuated towards MCF10A cells. B. Compared to MeWo cells, the cell toxicity of Cmpd-43 was significantly compromised to MEF cells. Supplemental Figure 3 Supplemental Figure 3. Both PRL2 and PRL3 could also form trimer, and Cmpd-43 could disrupt PRL2 and PRL3 trimerization. A. Amino acid sequence alignment of all PRLs. Residues involved in trimer interface were labeled as bold letters; while residues involved in Analog-3 binging were labeled as boxed letters. B. PRL2 and PRL3 trimerization in the presence of either Cmpd-43 or Analog-1 by in vitro crosslinking assay. Supplemental Figure 4 Supplemental Figure 4. Knocking down of both PRL1 and PRL2 significantly reduced both ERK1/2 and Akt activation in MeWo cells. Supplemental Figure 5 Supplemental Figure 5. Gross melanoma images after 20 days treatment of Cmpd-43. Supplemental Figure 6 Supplemental Figure 6. In vivo toxicity of Cmpd-43. A. 21 days of Cmpd-43 treatment (15 mg/kg or 30 mg/kg) showed no significant toxicity on major organs. B. The histological analysis indicates that 30 mg/kg of Cmpd-43 treatment for 20 days showed no toxicity.