* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download 3000_2013_1e
Minimal genome wikipedia , lookup
Metagenomics wikipedia , lookup
DNA barcoding wikipedia , lookup
Genome evolution wikipedia , lookup
Genome (book) wikipedia , lookup
Pathogenomics wikipedia , lookup
Gene expression profiling wikipedia , lookup
Gene expression programming wikipedia , lookup
Quantitative comparative linguistics wikipedia , lookup
Microevolution wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Koinophilia wikipedia , lookup
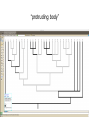
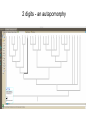
GENE 3000 Fall 2013 slides 127-159 wiki. wiki. wiki. 4 species can be related to each other in only 3 ways 4 species can be related to each other in only 3 ways B A C evolution - story of gain and loss • when and how were complex eyes evolved? in what species are they lost? are the genes required to develop eyes still there? can they be expressed in different ways? • • how frequently are genes gained and lost? how often is there lateral gene transfer between hosts and pathogens, or microbes? why are there more A/T bases in parts of the mitochondrion? Nature, June 2012 all starts with this • phylogeny: visual representation of the terms evolutionary history of populations, genes, species • tips: terminal ends of a phylogeny, representing the populations, genes, species • branches: lineages evolving through time • node: where lineages split; the common ancestor to descendants • clade: a group all descended from one • phylogeny: visual representation of the terms evolutionary history of populations, genes, species • tips: terminal ends of a phylogeny, representing the populations, genes, species • branches: lineages evolving through time • node: where lineages split; the common ancestor to descendants discovermagazine.com • clade: a group all descended from one organisms are related through descent from ancestors progression of generations more than 2 million identified and classified species extant taxonomy (systematics) • groups defined on previous slides are clades • group comprised of one organism and all its descendants (monophyly) • indication of homology Archosaurs how do we know? • inference, reconstruction • characters are heritable traits that can be compared (an A in position 117; body hair) • careful evaluation of homology • assume that having more things in common indicates more recent common ancestor (longer shared history) homology and homoplasy • similarity without shared descent is homoplasy - convergent evolution (wings) or evolutionary reversal (snakes don’t have legs) • homology may require full evidence, e.g. development of insect wings very different from development of bird wings phylogeny inference • • morphological characters (presence, absence, quality) DNA-based characters work the same (idea is there may be more independence, and more of them) our data matrix Text? in NEXUS format parsimony analysis • assume each state evolved only once (not necessarily true) • principle of parsimony: the simplest explanation is the most likely • we can identify how often this assumption is violated one character • tentacular forelimbs might be able to split into 2 clades, but cannot fully resolve tree parsimony is a CRITERION • our assumption of parsimony does not find the tree, it is a score given to a tree • trees are searched algorithmically for all possible topologies (or all possible better than score X) • a parsimony phylogeny analysis returns the set of most-parsimonious trees our tree eyeless “presence of 2 eyes” - obviously negatively correlated with “eyeless” tail digits dark stripes hmmm correlated with digits!? wings circular tail fin “forked” tail fin “protruding body” 2 digits - an autapomorphy our tree • parsimony tree(s) from class data matrix • which characters exhibit HOMOPLASY • remember: homoplasy may be evolutionary convergence/reversal, may also reflect our own uncertainty about the character and how it develops! • see Box 1 (chapter 4) for another example molecular data • we will come to this later how it is analyzed, but we see the problems of • • • • highly correlated, non-independent characters hard to identify very many characters thus not a fully resolved phylogeny (but good!) if you sequence a single gene region, might have 100 characters that are unbiased by the person collecting the data example of carnivores define “carnivore” please... why study phylogeny of carnivores? lets ask a simple question: did aquatic pinnipeds evolve only once, or were there multiple transitions to the sea? 12 characters, 10 carnivores synapomorphies 13 13 20 characters, 3 equally parsimonious trees our result is robust to this uncertainty: one origin of pinnipeds (consensus at left) parsimony • simple assumption, simple model - may lead to several ‘equally parsimonious’ results • more data will not always solve the problem • more complex models evaluate the non-independence of data, the empirical patterns of DNA substitution, and probability theory