* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download IBS Methods for Affected Pairs Linkage
Artificial gene synthesis wikipedia , lookup
Skewed X-inactivation wikipedia , lookup
Neuronal ceroid lipofuscinosis wikipedia , lookup
Tay–Sachs disease wikipedia , lookup
X-inactivation wikipedia , lookup
Fetal origins hypothesis wikipedia , lookup
Public health genomics wikipedia , lookup
Population genetics wikipedia , lookup
Neocentromere wikipedia , lookup
Medical genetics wikipedia , lookup
Genome-wide association study wikipedia , lookup
Microevolution wikipedia , lookup
Genetic drift wikipedia , lookup
Human leukocyte antigen wikipedia , lookup
Quantitative trait locus wikipedia , lookup
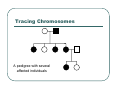
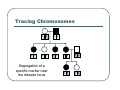
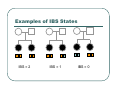
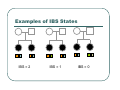
IBS Methods for Affected Pairs Linkage Biostatistics 666 Lecture 14 Genetic Mapping “Compares the inheritance pattern of a trait with the inheritance pattern of chromosomal regions” Positional Cloning “Allows one to find where a gene is, without knowing what it is.” Where are the genes influencing a particular trait? Intuition for Linkage Analysis z Millions of variations could potentially be involved • Costly to investigate each individually z Within families, variation is organized into a limited number of haplotypes • Sample modest number of markers to determine whether each stretch of chromosome is shared Tracing Chromosomes A pedigree with several affected individuals Tracing Chromosomes Segregation pattern for chromosome carrying disease alleles Tracing Chromosomes 1 2 1 3 1 4 Segregation of a specific marker near the disease locus 3 4 2 3 1 3 5 6 3 5 1 5 Tracing Chromosomes 12 12 12 14 14 14 13 13 13 34 34 34 13 14 14 Multiple nearby markers can segregate in a manner that tracks disease! 24 24 24 13 13 13 14 14 14 12 12 12 Today … z Linkage analysis with sibling pairs z Find markers that are near disease locus z Minimalist approach … • Identity-by-State (IBS) based method • Near means recombination fraction θ < ½ Reference for Today… z Power of IBS Methods for Linkage Analysis z Bishop DT and Williamson JA (1990) Am J Hum Genet 46:254-265 z Recommended Reading Bishop and Williamson (1990) Opening Line "The availability of a large number of DNA markers has made possible mapping projects with the certainty that if: (a) a major gene exists for a trait; (b) the trait is reasonably homogeneous; (c) there is sufficient family material available; then a linked marker can be found." Data for a Linkage Study z Pedigree z Observed marker genotypes z Phenotype data for individuals • Set of individuals of known relationship • SNPs, VNTRs, microsatellites Minimalist Approach z Pedigree z Observed Marker Genotypes z Phenotypes • Two individuals of known relationship • A single marker • Both individuals are affected Allele Sharing Analysis z z z Reject random sharing at a particular region Less powerful than classic methods • When disease model is known More robust than classic methods • When disease model is unknown Consider Autosomal Recessive Locus … z For a collection of sibling pairs… z What patterns of sharing do you expect at the disease locus? z What patterns of sharing to you expect as you move away from the disease locus? Historical References z Penrose (1953) suggested comparing IBD distributions for affected siblings. • Possible for highly informative markers (eg. HLA) z Thomson (1986) suggested discarding partially informative families. z Lange (1986) proposed using IBS information instead of IBD. IBS Based Methods z Sample of affected relative pairs z Examine a marker of interest z Count alleles shared for each pair • This includes both … • Chromosomes that are identical-by-descent • Chromosomes that simply carry identical alleles Examples of IBS States 1/ 1 1/1 IBS = 2 1/ 1 1/2 IBS = 1 2/ 2 1/1 IBS = 0 Examples of IBS States 1/ 2 1/2 IBS = 2 1/ 3 1/2 IBS = 1 1/ 2 3/4 IBS = 0 Evidence for Linkage z Increased similarity in affected pairs z Compared to: • Unselected pairs • Unaffected pairs • Discordant pairs • Expectations derived from allele frequencies Test for Independence χ 22df = ∑ i χ12df z z [N IBS =i − E ( N IBS =i )]2 (general test, for sibling pairs) E ( N IBS =i ) 2 2 [ N IBS =0 − E ( N IBS =0 )] [N IBS >0 − E ( N IBS >0 )] = + E ( N IBS =0 ) E ( N IBS >0 ) (grouping often preferable for other relatives) Assuming all counts are relatively large If counts are small, use binomial or trinomial distribution Modeling IBS Sharing z For any relative pair, calculate: • Probability of IBD sharing • 0, 1 or 2 alleles • Conditional probability of IBS sharing • 0, 1, 2 alleles • IBS sharing >= IBD sharing • Why? IBD z The underlying sharing of chromosomes segregating within a family z Siblings share 0, 1 or 2 alleles z • Probabilities ¼, ½ and ¼ Unilineal relatives share 0 or 1 alleles • Probability of sharing is kinship coefficient φ * 4 P(Marker Genotype|IBD State) Relative I II (a,b) (c,d) (a,a) (b,c) (a,a) (b,b) (a,b) (a,c) (a,a) (a,b) (a,b) (a,b) (a,a) (a,a) Prior Probability 0 4papbpcpd 2pa2pbpc pa2pb2 4pa2pbpc 2pa3pb 4pa2pb2 p a4 IBD 1 0 0 0 pa pb p c pa2pb (papb2+pa2pb) pa 3 2 0 0 0 0 0 2papb pa 2 ¼ ½ ¼ Note: Assuming alleles unordered within genotypes P(IBS = i | IBD = j) P( IBS = 2 | IBD = 0) = 2∑ pi2 p 2j + ∑ pi4 i≠ j i P( IBS = 1 | IBD = 0) = 4∑ pi2 p j (1 − pi − p j ) + 4∑ pi3 (1 − pi ) i≠ j i P( IBS = 0 | IBD = 0) = ∑ pi p j (1 − pi − p j ) 2 + ∑ pi2 (1 − pi ) 2 i≠ j i P(IBS = i | IBD = j) P ( IBS = 2 | IBD = 2) = 1 P( IBS = 1 | IBD = 2) = 0 P( IBS = 0 | IBD = 2) = 0 P( IBS = 2 | IBD = 1) = ∑ pi2 i P( IBS = 1 | IBD = 1) = ∑ pi p j i≠ j P( IBS = 0 | IBD = 1) = 0 Example, Assuming Equal Allele Frequencies P(IBS=0) P(IBS=1) P(IBS=2) 2 alleles, IBD=0 .125 .500 .375 2 alleles, IBD=1 .000 .500 .500 3 alleles, IBD=0 .222 .592 .185 3 alleles, IBD=1 .000 .666 .333 Inference from Example z IBS approaches IBD as number of alleles increases z If linkage is being tested with chi-square test, how does the number of alleles (and marker informativeness) affect these two tests: • A test of whether NIBS >= 1 increases? • A test of whether NIBS > 1 increases? IBS Probabilities No. of Alleles P(IBS=0) P(IBS=1) P(IBS=2) 2 .03 .37 .60 3 .05 .48 .47 4 .08 .51 .40 20 .21 .52 .27 ∞ .25 .50 .25 Sibling IBS as a function of allele count, for marker with equally frequent alleles Results of Bishop and Williamson (1990) z Effect size, P(IBS | Affected pair) z Number of alleles at marker z Different relationships z Recombination fraction More Alleles Increase Power Effect of Recombination Varies According to Relationship Power vs. P(IBS | Affected Pair) With no phenocopies, rare alleles are easier to map In general, phenocopies decrease power Shortcomings of IBS Method z All sharing is weighted equally z Inefficient. • Sharing a rare allele • Sharing a common allele • Sharing homozygous genotype • Sharing heterozygous genotype Recommended Reading z Bishop DT and Williamson JA (1990) Am J Hum Genet 46:254-265 z Good introduction to linkage analysis in affected relative pairs, discusses • • • • Marker choice Recombination fraction Disease model Type of relative pair