* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Genetic approaches to development: Drosophila as a model organism
Microevolution wikipedia , lookup
Gene expression profiling wikipedia , lookup
Oncogenomics wikipedia , lookup
Primary transcript wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Epigenetics in stem-cell differentiation wikipedia , lookup
Genome (book) wikipedia , lookup
Epigenetics of human development wikipedia , lookup
X-inactivation wikipedia , lookup
Designer baby wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Mir-92 microRNA precursor family wikipedia , lookup
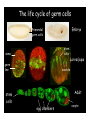
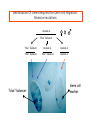
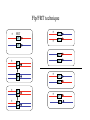
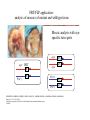
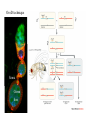
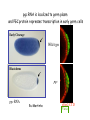
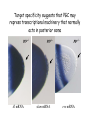
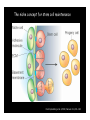
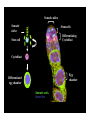
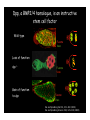
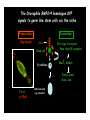
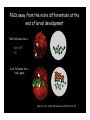
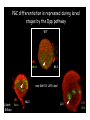
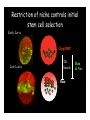
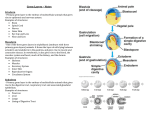
Genetic approaches to development: Drosophila as a model organism Ruth Lehmann New York University/Howard Hughes Medical Institute [email protected] Model Organisms and Innovative Approaches in Developmental Biology, Juquehy, Brazil-2005 Lecture: Genetic Analysis in Drosophila Ruth Lehmann [email protected] Embryogenesis Gastrulation Generation time Life span Eggs/female Fertilization # of autosomes Sex chromosomes LIFE CYCLE STATISTICS C. elegans Drosophila 14 hrs 24 hrs 2 hrs 3 hrs 3 days 10 days 20 days 2-3 months 300 (+) ~700 Internal Internal 5 3 XX, XO XX, XY zebrafish 48 hrs 6 hrs 3 month > 3 years ~15 000 external 25 none Species Chromosomes CM E. coli S. cerevisiae C. elegans D. melangaster D. rerio M.musculus H. sapiens 1 16 6 4 25 20 23 N/A 4000 300 280 2900 1700 3300 DNA content/haploid genome inMB 5 12 100 165 1740 3000 3000 Year sequence complete 1997 1997 1998 2000 2004 2003 2001 Genes/haploid 4,000 6,000 19,000 14,000 40,000 30,000 30,000 A brief history of fly genetics •1910 Morgan identifies white •1930 Calvin Bridges linkage groups and polytene chromosomes •1930 Sturtevant clonal analysis •1948 Balancer Chromosomes •1968 Lewis and Bacher EMS •1934 to 1965 From 572 stocks in 1934 to 15 000 in 1965 •1980 Systematic mutant screens for embryonic lethals by Nüsslein-Volhard and Eric Wieschaus •1980 P element techniques by Rubin and Spradling •2000 Genome sequenced •2005 Mutations in 7000 genes deletions for most of the genome Gonadcycle development The life of germ cells Primordial germ cells Embryo TF Embryo soma stem cells germ line ovariole Larva/pupa Adult stem cells egg chambers oocyte Why flies???? Genetics Different Types of Mutageneses Mutagenic Effect Advantages Disadvantages EMS (and other chemicals) Base pair changes (point mutations) Random Saturation Different types of mutations Slow gene identification Large-scale P-elements (and other transposons) DNA Insertions (mostly hypomorphic) Fast gene identification Flexible scale No saturation Non-random (hotspots) Deficiency kit (and other aberrations) Chromosome rearrangements (gene deletions) Small-scale Fast screening Defined set Slow gene identification No real saturation Ionizing radiations (Xrays, g-rays) Chromosome rearrangements (gene deletions) Random Gene deletions Slow gene identification No real saturation Inefficient Forward Genetics in Drosophila any mutagen -Zygotic screen (e.g. Wieschaus & Nüsslein-Volhard) -Maternal-effect screen (e.g. Schüpbach & Wieschaus) -Maternal-effect clonal screen (e.g. Perrimon, St Johnston) -Adult clonal screen (e.g. Dickson) -Modifier (Enhancer/Suppressor) screen (e.g. Rubin) Assays: You can only find what you are looking for Primary and secondary screens •Lethality •Sterility •Behavior •Pattern defects: segmentation, eye •Gene expression: RNA, protein, lacZ, GFP General mutagenesis approach to isolate zygotic genes EMS DTS Bal P X RT F1 single DTS Bal *Mutagenized chromosome* Bal *Mutagenized chromosome* Bal F2 F3 X *Mutagenized chromosome* Bal *Mutagenized chromosome* *Mutagenized chromosome* Test phenotype X Bal Bal Identification Of Genes Required For Germ Cell Migration: Recessive mutations mutant A ‘blue’ balancer ‘blue’ balancer mutant A mutant A ‘blue’ balancer ‘blue’ balancer mutant A “blue”-balancer Germ cell marker Drosophila germ cells from in germ plasm that assembles at the posterior pole during oogenesis Germ plasm Germ cells General mutagenesis approach to isolate mutations in maternal effect genes EMS DTS Bal P X RT F1 single DTS Bal X *Mutagenized chromosome* Bal RT F2 F3 F4 *Mutagenized chromosome* Bal *Mutagenized chromosome* *Mutagenized chromosome* Bal *Mutagenized chromosome* progeny X Bal Bal Test phenotype The “No Germ Cells” Class wild-type “no germ cells” How to identify all genes in a process? I. Same gene plays role during many stages/in many tissues Flp/FRT technique * * * * FRT > > * * > > > > > > > > * > > > > * > > > > FRT/FLP application: analysis of mosaics of mutant and wildtype tissue Mosaic analysis with eyespecific twin spots agoago- FRT P(w+) > > agoP(w+) P(w+) > > > > KENNETH H. MOBERG, DAPHNE W. BELL, DOKE C. R. WAHRER, DANIEL A. HABER & ISWAR K. HARIHARAN Nature 413, 311 - 316 (2001); Archipelago regulates Cyclin E levels in Drosophila and is mutated in human cancer cell lines OvoD technique Soma Germ line FRT/FLP application: Lineage analysis +/wt +/- hs-flp ; hs FRT FRT +/+ -/- FRT FRT, nls-GFP -/- FRT, nls-GFP FRT, nls-GFP +/- or +/+ Enhancer/suppressor screens Sensitized condition: @ 22.7oC sevts @ 24.3oC R7 present R7 absent a ts mutation in kinase domain EMS P F1 sev- ; + ; sev+ or Bal, P(sevts) D sevD2 ; + ; + X Y + + sev- ; * ; * sev-/Y + Bal, P(sevts) Screen for absence of R7 at 22.7oC Different Mapping Methods in Drosophila Method tools principle result resolution pro con Classical meiotic mapping Based on visible markers Meiotic Genetic recombination map position Non-molecular (not all markers cloned) genetic location Few visible markers available, often spaced far away from mutant Deficiency mapping Based on available deficiencies Complementation Chromosomal interval Non-molecular (not all breakpoints molecularly mapped) Fast mapping to a certain region Not all regions of the genome covered Interactions with other genes in Df P-mediated male recombination mapping Based on available P insertions Homologous recombination Position of mutation relative to P (proximal/distal) Molecular (if position of P known) Precise molecular interval Stepwise process, slow Still requires visible markers SNP mapping Based on molecular markers Meiotic Molecular map position Molecular Markers neutral Precise molecular interval Expensive dependent on detection method Forward Genetics in Drosophila any mutagen -Zygotic screen (e.g. Wieschaus & Nüsslein-Volhard) -Maternal-effect screen (e.g. Schüpbach & Wieschaus) -Maternal-effect clonal screen (e.g. Perrimon, St Johnston) -Adult clonal screen (e.g. Dickson) -Modifier (Enhancer/Suppressor) screen (e.g. Rubin) P-element based -Enhancer-trap screen (e.g. Bellen, Jan) -Overexpression-trap screen (e.g. Rorth) -Protein-trap screen (e.g. Chia, Cooley) Venken & Bellen, March 2005 Mis/overexpression screens, the good and the bad wrong gene --- right pathway Faf-lacZ; Gal4-driver X nos nos5’UTR Gal4-VP16 nos3’UTR EP-UAS-insertion lines UAS Endogenous gene Expression in germ cells “mes” Gal4 UAS Endogenous gene Expression in soma st 14 dorsal a-Vasa wild type over- or mis- expression These screens can be misleading-gene is not expressed in germ cells and has no phenotype in germ cells st 9 tre1 hemocytes st 13 caudal visceral mesoderm midgut primordia midgut glia ..but RNA of close homolog is localized to the germ plasm and PGCs tre1 RNA Stage 3 and mutations in this gene affect PGCs migration Stage 13 Prabhat Kunwar Mis/overexpression screens, the good and the bad “Redundant” genes wunen RNA wunen 2 RNA Zhang et al, Nature (1997) 385, 64-67; Starz-Gaiano et al, Development (2001) 128, 983-991 Mis/overexpression screens can identify “redundant” genes wun-/- and wun2-/- of both genes mes::Gal4; UAS::wun2 Stage 11 either gene Vasa Zhang et al. (1997) Nature 385, 64-67 Starz-Gaiano et al. (2001) Development 128, 983-991 How to identify all genes in a process? II. Technologies beyond EMS and Pelements Reverse Genetics in Drosophila -Dominant negative (GAL4-UAS based) -RNAi (injection, GAL4-UAS based) -Homologous recombination -Tilling Keep balanced stock Venken & Bellen, March 2005 Venken & Bellen, March 2005 Knowing when to Stop Screening: Efficiency and Saturation Wieschaus and Nüsslein-Volhard Germ Cells •Set aside early in development from somatic cells •Highly specialized (migration, cell interaction, meiosis) •The ultimate stem cell, able to generate new generation •The ultimate stem cell, able to generate new generation Egg & Sperm Zygote Germ line stem cells Primordial Germ Cells Soma Early segregation protects germ cells from somatic differentiation Death Germ cells are specificied by maternally synthesized germ plasm or by cell-to cell induction Germplasm Drosophila, Xenopus, zebrafish, C. elegans Induction Mouse, axolotl Genetically distinct pathways control formation of germ line and soma in the early in Drosophila emrbyo Nuclear migration Budding Polarized membrane Growth Germ cells Somatic cells Fly and mouse germ cells: repression of somatic genes Transcription is repressed in early germ cells blue: slam RNA green: Vasa red: pSer2-CTD Stein et al. (2002) Development 129(16), 3925-3934 Seydoux & Dunn (1997) Development 124(11), 2191-2201 pgc RNA is localized to germ plasm and PGC protein represses transcription in early germ cells Early Cleavage Wild-type Blastoderm pgc pgc RNA Rui Martinho pSer2-CTD Vasa slam and other “somatic genes”are activated in pgc mutant germ cells Wild type pgc Wild type pgc as-pgc slam RNA Rui Martinho tailless mRNA Martinho et al. ( 2004) Current Biol. 14(2), 159-165 PGC may repress germ cell transcription by interfering with transcriptional elongation CTD phosphorylation recruits Set1 and Set2 histone methylases How are germ cells set aside from somatic cells Soma Germ Cells Somatic signals pgc Somatic differentiation Target specificity suggests that PGC may repress transcriptional machinery that normally acts in posterior soma pgc-/- tll mRNA pgc-/- slam mRNA pgc-/- eve mRNA Cross-regulation of torso and pgc pathways may inhibit cell specification of soma vs germ cells Soma Germ Cells Torso pgc Receptor tyrosine kinase Somatic target genes Germ cell target genes Up-regulation of torso represses germ cell formation Wild-type (100%) 8 copies torso+ (35%) (25%) Rui Martinho Up-regulation of pgc leads to somatic cellularisation defects similar to the ones observed in torso loss of function alleles Wild-type green: Vasa 6 copies pgc+ blue: DNA torLOF Rui Martinho Antagonism between pgc and torso sets apart somatic cells from germ cells Soma Germ Cells torso pgc Somatic target genes Germ cell target genes Posterior soma Germ cells Repression of somatic differentiation via transcriptional regulation could be critical for germ cell specification Germ cell specification in Drosophila slam RNA (somatic gene) Primordial germ cells Germ cell specification in mice (Saitou et al., 2002) Repression of somatic differentiation via transcriptional regulation a common theme for germ cell specification? Germ cell specification in Drosophila Germ cell specification in mice (Saitou et al., 2002) Hoxb1 (somatic gene) slam RNA (somatic gene) fragilis (germ line marker) Primordial germ cells Primordial germ cells Fly and mouse germ cells: pgc = stem cells? The niche concept for stem cell maintenance From:Spradling et al. (2001) Nature 414, 98-104 Somatic niche Somatic niche Stem cells Differentiating Cystoblast Stem cell Cystoblast Egg chamber Differentiated egg chamber Somatic cells Germ line Dpp, a BMP2/4 homologue, is an instructive stem cell factor Wild type Fusome Vasa Loss of function dpp-/- Fusome Vasa Gain of function hs-dpp Fusome Vasa Xie and Spradling Cell 94, 251-260 (1998) Xie and Spradling Science 290, 328-330 (2000) The Drosophila BMP2/4 homologue DPP signals to germ line stem cells via the niche Soma/niche Dpp ligand Germ line Niche Stem cell Cystoblast Tkv (type I receptor) Punt (type II receptor) MadP, Medea Target genes Bam, dad Vasa p-Mad Differentiated egg chamber The number of germ cells increases dramatically during larval stages EE 250 LL3 150 Number of adult GSCs/Ovary 50 24 h 48 h 72 h 96 h Hours after egg laying ~108 h Bar: 20 mm PGCs away from the niche differentiate at the end of larval development Mid 3rd instar larva bam::GFP 1B1 Bam-GFP hts Late 3rd instar larva /early pupa Early pupa Zhu CH, Xie T. (2003) Development,130(12):2579-88. PGC differentiation is repressed during larval stages by the Dpp pathway WT 1B1 pMad ML3 nos-Gal4 X UAS-dad Lilach Gilboa 1B1 Vasa ML3 LL3 1B1 Orb Restriction of niche controls initial stem cell selection Early Larva Dpp/BMP Late Larva Tkv Smads Bam Pum & Nos Primordial germ cell = germ line stem cell? Vasa pSer2-CTD 1B1 Vasa Niki Y, Mahowald AP. (2003) Proc Natl Acad Sci U S A; 100(24):14042-5 Gilboa L, Lehmann R. (2004) Curr Biol;14(11):981-6. Wang Z, Lin H. (2004) Science; 303(5666):2016-9. Epub 2004 Feb 19.