* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download MCB Test 3 Review
G protein–coupled receptor wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Secreted frizzled-related protein 1 wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Metalloprotein wikipedia , lookup
Point mutation wikipedia , lookup
Polyclonal B cell response wikipedia , lookup
Expression vector wikipedia , lookup
Signal transduction wikipedia , lookup
Biochemical cascade wikipedia , lookup
Interactome wikipedia , lookup
Western blot wikipedia , lookup
Paracrine signalling wikipedia , lookup
Protein purification wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Homology modeling wikipedia , lookup
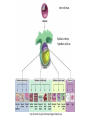
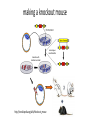
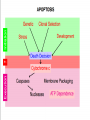
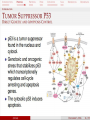
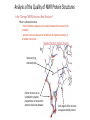
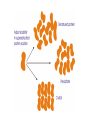
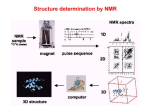
MCB Test 3 Review M. Alex Miranda 12/17/16 Stem Cell Biology Jim Huettner 11/22/2016 Stem Cells: definition • Self Renewal - undifferentiated cells that can divide repeatedly while maintaining their undifferentiated state. • Pluripotency – ability to differentiate into a variety of different cell types Types of Stem Cells Embryonic – from the inner cell mass of preimplantation embryos, prior to formation of the 3 germ layers (ectoderm, mesoderm, endoderm) Somatic – undifferentiated cells found in specific locations in “mature” tissues iPS cells – induced pluripotent stem cells generated by reprogramming differentiated cells (or cell nuclei, i.e. therapeutic cloning) Potency • Totipotent – able to generate every cell type including extraembryonic tissues • Pluripotent – able to generate cells from all three embryonic germ layers • Multipotent – able to generate a variety of cells from a particular somatic structure • Unipotent – only generate one cell type Inner cell mass Epiblast: embryo Hypoblast: yolk sac http://stemcells.nih.gov/info/scireport/pages/chapter1.aspx making a knockout mouse http://en.wikipedia.org/wiki/Knockout_mouse Pluripotency markers • Stage-specific antigens: Anti-SSEA 3 and 4 recognize globo-series gangliosides • Tra1-60 and Tra1-81: keratin sulfate surface antigens • Oct3/4, Sox2, Nanog – transcription factors involved with maintaining pluripotency • Normal karyotype, and pre-X-inactivation? Two types of ES cells? “Naïve” (ICM-like) • • • • • • Blastocyst chimera (+) High cloning efficiency Short doubling time Xa Xa Distal Oct4 enhancer High Nanog, Klf2/4, Rex1 “Primed” (Epi-SC) • • • • • • Blastocyst chimera (-) Low cloning efficiency Long doubling time Xa Xi Proximal Oct4 enhancer Low Nanog, Klf2/4, Rex1 Both types can self renew and give rise to cells from all 3 germ layers in teratomas or following in vitro differentiation maintenance of pluripotency - 2 “naïve” • • • • Positive Regulators LIF - Stat3 BMP4 - Smad1/5 Wnt (GSK-3 inhibitors) IGF • • • • • “primed” TGFb/activin – Smad2/3 FGF2 ERK1/2 Wnt (GSK-3 inhibitors) IGF Negative Regulators • TGFb/activin-Smad2/3 • FGF2 • ERK1/2 • BMP4 – Smad1/5 Reprogramming • SCNT – somatic cell nuclear transfer (reproductive and therapeutic cloning) – deterministic and fairly rapid • iPS – induced pluripotent stem cells – slow and stochastic (until recently) • Transdifferentiation – conversion of one terminally differentiated cell type into another without dedifferentiation to an immature phenotype. Must rule out cell fusion or other explanations. Reprogramming: somatic cell nuclear transfer http://www.biotechnologyonline.gov.au/images/contentpages/scnt.gif Generating iPS cells • Express transcription factors: Oct3/4, Sox2, Klf4 and c-Myc OR Oct3/4, Sox2, Nanog and Lin28 • Initial de-differentiation and proliferation (day 1-3, enhanced by Myc); histone modification and chromatin reorganization • 2nd wave of gene expression - stem cell and development related genes (day 9-12); DNA demethylation and X reactivation Removing the bottle neck? • Rais et al., Nature 502:65-70, 2013 implicate Mbd3, a component in the NuRD complex that mediates gene repression via histone deacetylation and chromatin remodeling. • Argue that the reprogramming factors recruit both repressive (Mbd3/NuRD) and de-repressive (Wdr5 and Utx) complexes, and reprogramming only occurs when the Mbd3/NuRd repression loses. • Achieve nearly 100% reprogramming within 7 days in cells with Mbd3 reduced or eliminated. Transdifferentiation • Conversion from one differentiated cell type to another without evident de-differentiation and re-differentiation • Must not be confused by cell fusion or selection for rare pluripotent cells in the source material. • Induced by expression of transcription factors and microRNAs Intracellular Protein Degradation Chris Weihl MD/PhD [email protected] Department of Neurology Consequence of impaired protein degradation • Protein aggregates • Ubiquitinated inclusions • Vacuolation • Damaged organelles • Secondary impairment in other cellular processes • Cell Death • Underlying pathogenesis of degenerative disorders (neurodegeneration, muscle degeneration, liver degeneration, lung disease, aging) Protein Degradation in the Cell Ub Autophagy Nucleus Aggresome Ub UPS Ub Ub Endocytosis Protein Degradation Ubiquitin/Proteasome Pathway 80-90% Most intracellular proteins • Lysosomal processes 10-20% Extracellular proteins Cell organelles Some intracellular proteins Ubiquitination of proteins is a FOUR-step process First, Ubiquitin is activated by forming a link to “enzyme 1” (E1). Then, ubiquitin is transferred to one of several types of “enzyme 2” (E2). Then, “enzyme 3” (E3) catalizes the transfer of ubiquitin from E2 to a Lys e-amino group of the “condemned” protein. Lastly, molecules of Ubiquitin are commonly conjugated to the protein to be degraded by E3s & E4s AMP PROTEASOME COMPONENTS 20S Proteasome 19S Particle ATP 26S Proteasome DEUBIQUITINATION De-ubiquitinating Autophagy • Lysosomal degradation of proteins and organelles • Occurs via three routes • Macroautophagy • Microautophagy (direct uptake of cellular debris via the lysosome) • Chaperone mediated autophagy (selective import of substrates via Hsc70 and Lamp2a) Selective Autophagy • Aggregaphagy– p62/SQSTM1, Nbr1 • Mitophagy – Parkin, Nix • Reticulophagy – endoplasmic reticulum • Ribophagy – translating ribosomes • Xenophagy – e.g. Salmonella via optineurin • Lipophagy – autophagy mediated lipolysis • Performed by an expanding group of ubiquitin adaptors Rapamycin as an inducer of autophagy Immunosuppressant used to treat transplant rejection Inhibits the mTOR pathway mTOR integrates extrinsic growth signals and cellular nutrient status and energy state Active mTOR Protein synthesis and cell growth Inactive mTOR (or rapamycin treatment) Inhibition of protein synthesis and increased autophagic degradation of protein TITLE P AGE I NTRODUCTION THE P ROCESS H IS TORIC AL L A N D M A R K S N O N L I N E A R D EVEL O PM E N TAL P ROGRAM AND S IGNALING P ATHWAYS BCL-2 P ROTEINS P ORE F ORMING S TRUCTURE T HE C ELL B IOLOGY OF A POPTOSIS T O L IVE IS TO DIE – METALLICA (2007) Paul H. Schlesinger Department of Cell Biology and Physiology Office McDonnell 401 Washington University Medical School [email protected] December 5, 2016 S CHLESINGER ( WUMS ) A POPTO D ECEMBER 9, 2014 I NITIATIN TITLE P AGE I NTRODUCTION C LAS S IFIC ATION OF THE P ROCESS S IGNALING P ATHWAYS BCL-2 P ROTEINS P ORE F ORMING S TRUCTURE I NITIATIN C ELLULAR D EATH H OW C ELLS A CHIEVE M ORTALITY Apoptosis – Programmed cell death, controlled part of development Necrosis – Premature cell death caused by external factors Autophagy – Degradation of cell components in lysosome. Type of programmed cell death, separate from apoptosis Senescence – Cell cycle arrest S CHLESINGER ( WUMS ) A PO PTO SIS D ECEMBER 9, 2014 3 / 37 TITLE P AGE I NTRODUCTION THE P ROCESS S IGNALING P ATHWAYS BCL-2 P ROTEINS P ORE F ORMING S TRUCTURE I NITIATIN MORPHOLOGICAL A POPTOSIS : M ORPHOLOGY Morphological Progression Retain Membrane Barriers S CHLESINGER ( WUMS ) A PO PTO SIS D ECEMBER 9, 2014 9 / 37 NMR in biology: Structure, dynamics and energetics Gaya Amarasinghe, Ph.D. Department of Pathology and Immunology [email protected] CSRB 7752 • Structure determination by NMR • NMR relaxation– how to look at molecular motion (dynamics by NMR) • Ligand binding by NMR – Energetics Protein Structures from an NMR Perspective Background – We are using NMR Information to “FOLD” the Protein. – We need to know how this NMR data relates to a protein structure. – We need to know the specific details of properly folded protein structures to verify the accuracy of our own structures. – We need to know how to determine what NMR experiments are required. – We need to know how to use the NMR data to calculate a protein structure. – We need to know how to use the protein structure to understand biological function Protein Structures from an NMR Perspective Analyzing NMR Data is a Non-Trivial Task! there is an abundance of data that needs to be interpreted X Interpreting NMR Data Requires Making Informed “Guesses” to Move Toward the “Correct” Fold Distance from Correct Structure Not A Direct Path! Initial rapid convergence to approximate correct fold Correct structure NMR Data Analysis Iterative “guesses” allow “correct” fold to emerge Nuclei are positively charged many have a spin associated with them. Moving charge—produces a magnetic field that has a magnetic moment Spin angular moment Why use NMR ? Some proteins do not crystallize (unstructured, multidomain) crystals do not diffract well can not solve the phase problem Functional differences in crystal vs in solution can get information about dynamics Protein Structure Determination by NMR •Stage I—Sequence specific resonance assignment •State II – Conformational restraints •Stage III – Calculate and refine structure Protein Structure Determination by NMR •Stage I—Sequence specific resonance assignment •State II – Conformational restraints •Stage III – Calculate and refine structure NMR Structure Determination NOE NOE - a through space correlation (<5Å) - distance constraint 4.1Å 2.9Å Coupling Constant (J) - through bond correlation J NH CaH - dihedral angle constraint Chemical Shift - very sensitive to local changes in environment - dihedral angle constraint Dipolar coupling constants (D) - bond vector orientation relative to magnetic field - alignment with bicelles or viruses CaH D NH Protein Structure Determination by NMR •Stage I—Sequence specific resonance assignment •State II – Conformational restraints •Stage III – Calculate and refine structure Protein Structures from an NMR Perspective What Information Do We Know at the Start of Determining A Protein Structure By NMR? Effectively Everything We have Discussed to this Point! The primary amino acid sequence of the protein of interest. ► All the known properties and geometry associated with each amino acid and peptide bond within the protein. ► General NMR data and trends for the unstructured (random coiled) amino acids in the protein. The number and location of disulphide bonds. ► Not Necessary can be deduced from structure. Analysis of the Quality of NMR Protein Structures With A Structure Calculated From Your NMR Data, How Do You Determine the Accuracy and Quality of the Structure? • Consistency with Known Protein Structural Parameters bond lengths, bond angles, dihedral angles, VDW interactions, etc all the structural details discussed at length in the beginning • Consistency with the Experimental DATA distance constraints, dihedral constraints, RDCs, chemical shifts, coupling constants all the data used to calculate the structure • Consistency Between Multiple Structures Calculated with the Same Experimental DATA Overlay of 30 NMR Structures Analysis of the Quality of NMR Protein Structures Is the “Average” NMR Structure a Real Structure? • No-it is a distorted structure level of distortions depends on the similarity between the structures in the ensemble provides a means to measure the variability in atom positions between an ensemble of structures Expanded View of an “Average” Structure Some very long, stretched bonds Position of atoms are so scrambled the graphics program does not know which atoms to draw bonds between Some regions of the structure can appear relatively normal Protein crystallography in practice MCB 15 Dec 2016 Daved H. Fremont [email protected] Department of Pathology and Immunology Washington University School of Medicine An 7-step program for protein structure determination by x-ray crystallography 1. Produce monodisperse protein either alone or as relevant complexes 2. Grow and characterize crystals 3. Collect X-ray diffraction data 4. Solve the phase problem either experimentally or computationally 5. Build and refine an atomic model using the electron density map 6. Validation: How do you know if a crystal structure is right? 7. Develop structure-based hypothesis 1. Produce monodisperse protein either alone or as relevant complexes Methods to determine protein purity, heterogeneity, and monodispersity Gel electrophoresis (native, isoelectric focusing, and SDS-PAGE) Size exclusion chromatography Dynamic light scattering http://www.protein-solutions.com/ Circular Dichroism Spectroscopy http://www-structure.llnl.gov/cd/cdtutorial.htm Characterize your protein using a number of biophysical methods Establish the binding stoichiometry of interacting partners 2. Grow and characterize crystals Hanging Drop vapor diffusion Sitting drop, dialysis, or under oil Macro-seeding or micro-seeding Sparse matrix screening methods Random thinking processes, talisman, and luck The optimum conditions for crystal nucleation are not necessarily the optimum for diffraction-quality crystal growth Space Group P21 4 M3 /ASU diffraction >2.3Å 14.4% Peg6K NaCacodylate pH 7.0 200mM CaCl2 Space Group C2 2 M3 /ASU diffraction >2.1Å 18% Peg4K Malic Acid/Imidazole pH 5.1 100mM CaCl2 Hanging Drop Sitting drop Commercial screening kits available from http://www.hamptonresearch.com; http://www.emeraldbiostructures.com Space Group P3121 3 M3 + 3 MCP-1/ASU diffraction > 2.3Å 18% Peg4K NaAcetate pH 4.1 100mM MgCl2 3. Collect X-ray diffraction data Initiate experiments using home-source x-ray generator and detector Determine liquid nitrogen cryo-protection conditions to reduce crystal decay While home x-rays are sufficient for some questions, synchrotron radiation is preferred Anywhere from one to hundreds of crystals and diffraction experiments may be required Argonne National Laboratory Structural Biology Center beamlineID19 at the Advanced Photon Source http://www.sbc.anl.gov 4. Solve the phase problem either experimentally or computationally Structure factor equation: By Fourier transform we can obtain the electron density. We know the structure factor amplitudes after successful data collection. Unfortunately, conventional x-ray diffraction doesn’t allow for direct phase measurement. This is know as the crystallographic phase problem. Luckily, there are a few tricks that can be used to obtain estimates of the phase a(h,k,l) Experimental Phasing Methods MIR - multiple isomorphous replacement - need heavy atom incorporation MAD - multiple anomalous dispersion- typically done with SeMet replacement MIRAS - multiple isomorphous replacement with anomalous signal SIRAS - single isomorphous replacement with anomalous signal Computational Methods MR - molecular replacement - need related structure Direct and Ab Initio methods - not yet useful for most protein crystals 5. Build an atomic model using the electron density map Electron density for the AP-2 aappendage Initial bones trace for the AP-2 aappendage Final trace for the AP-2 aappendage Low-resolution At 4-6Å resolution, alpha helices look like sausages. Medium resolution ~3Å data is good enough to see the backbone with space in between. Holes in rings are a good thing Seeing a hole in a tyrosine or phenylalanine ring is universally accepted as proof of good phases. You need at least 2Å data. 6. Validation: How do you know if a crystal structure is right? The R-factor R = S(|Fo-Fc|)/S(Fo) where Fo is the observed structure factor amplitude and Fc is calculated using the atomic model. R-free An unbiased, cross-validation of the R-factor. The R-free value is calculated with typically 5-10% of the observed reflections which are set aside from atomic refinement calculations. Main-chain torsions: the Ramachandran plot Geometric Distortions in bond lengths and angles Favorable van der Waals packing interactions Chemical environment of individual amino acids Location of insertion and deletion positions in related sequences 6. Validation: Mapping of sequence conservation in AP-2 a-subunit appendages Traub LM, Downs MA, Westrich JL, and Fremont DH: (1999) Crystal structure of the a-appendage of AP-2 reveals a recruitment platform for clathrin-coat assembly. Proc. Natl. Acad. Sci. U.S.A. 96:8907-8912. 7. Develop structure-based hypothesis Structure-Based Mutagenesis of the a-appendage Traub LM, Downs MA, Westrich JL, and Fremont DH: (1999) Crystal structure of the a-appendage of AP-2 reveals a recruitment platform for clathrin-coat assembly. Proc. Natl. Acad. Sci. U.S.A. 96:8907-8912.