* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Repair of DNA double-strand breaks and susceptibility to breast
Behavioural genetics wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Population genetics wikipedia , lookup
Human genetic variation wikipedia , lookup
Public health genomics wikipedia , lookup
Microevolution wikipedia , lookup
Genome-wide association study wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Oncogenomics wikipedia , lookup
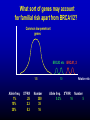
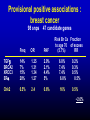
Breast/Ovarian Family † 57 54 22 43 † 49 55 48 51 32 + + PO 45 PO 45 B 32 O 59 39 Inherited predisposition More BRCA-like genes Rare, moderately strong variants Common genetic variation Role of normal genetic variation in determining individual risk. How useful is this information in selection for screening and prevention? How do we find the genes? Breast cancer as an example Evidence that genetic variation affects risk Measure of variation = familial clustering Risk in close blood relative compared to risk in population as a whole = roughly 2-fold. Is family clustering genetic? MZ twin DZ twin Mother/sister Incidence % per year 1.31 0.5 0.36 Patient’s contralateral breast 0.66 (Peto & Mack, Nat Genet 26, 411 (2000)) How much genetic predisposition is there? How is it distributed? Determines potential for discriminating individual risks risk Breast/Ovarian Family † 57 54 22 43 † 49 55 48 51 32 + + PO 45 PO 45 B 32 O 59 39 Familial clustering of breast cancer OBS EXP Excess 177 106 71 Population 13 1.47 11.5 BRCA1/2 mutation Fraction of excess familial clustering attributable to BRCA1/2 = 15-20% Familial clustering of breast cancer Risk to 1o relative of case Roughly 15-20% due to BRCA1/2 2 Excess familial risk 1 ATM Chk-2 Ha-ras PTEN What sort of genes may account for familial risk apart from BRCA1/2? Common low-penetrant genes BRCA3 etc 1.5 Allele freq. 1% 10% 30% XsFRR .25 2.3 5.3 Number 350 35 16 10 Allele freq. 0.2% BRCA1, 2 Relative risk XsFRR Number 16 5 Patterns of breast cancer in families 1500 cases, population based BRCA1/2 excluded What model fits best? Best fit = combined result of several factors, individually of small effect = log-normal distribution of risk in population. Distribution of genotypes in population and cases by genotype risk 0.040 SD = 1.2 0.030 Population Cases 0.020 0.010 0.000 0.01 0.10 1.00 Relative risk 10.00 100.00 Proportion of population and cases above specified risk: SD = 1.2 Proportion above given risk (x) 100% 88% Cases Population 50% 46% 10% 0% 0% 3% 12% 20% 40% 60% Risk of breast cancer by age 70 80% Effects of normal genetic variation on breast cancer risks Population 10% Cancers Individual risk by age 70 50% 46% 12% >1:8 < 1 : 30 Proportion of population and cases above specified risk: SD = 0.8 Proportion above given risk (x) 100% 80% Cases Population 50% 31% 10% 0% 0% 4% 11% 20% 40% Risk of breast cancer by age 70 60% 80% Proportion of population and cases above specified risk: SD = 0.3 Proportion above given risk (x) 100% Cases Population 75% 50% 25% 0% 0% 20% 40% Risk of breast cancer by age 70 60% 80% Gail model of breast cancer risk Nurses Health Study Analysis Excellent prediction of breast cancer incidence in specified population. Poor prediction of risk to individual. 2.8-fold between upper and lower deciles cut-off for tamoxifen use defined 33% of population with 44% of cases. (Rockhill, JNCI 93, 358 (2001)) - find genes - interactions - validation 1/5 1/5 40x risk QuickTime™ and a Photo - JPEG decompressor are needed to see this picture. How to find the genes? Association studies C T arg cys V indirect direct linkage disequilibrium Problems: recombination origins different time multiple origins Common variant : common disease Marker Disease allele Rare variants Candidate genes Estrogen synthesis and degradation; ER Cell cycle checkpoints DNA repair TGFb pathway IGF pathway Carcinogen metabolism Sample sets Initial : 2000 cases, 2000 controls Confirmatory : 2000 cases, 2000 controls Cases - Population based, East Anglia simple epidemiology data, survival; paraffin blocks Controls - EPIC cohort, East Anglia extensive epidemiological data, follow-up, serum, mammography, bone density, etc Power Percentage polygenic variance explained. 90% power p = 10-4 multiplicative 6000 Sample size 5000 4000 1% 2% 3000 5% 2000 10% 1000 0 0 0.05 0.1 0.15 0.2 0.25 0.3 0.35 Allele Frequency (Antoniou & Easton, submitted) Provisional positive associations : breast cancer 98 snps 47 candidate genes Risk Br Ca Fraction to age 70 of excess (5.7%) RR Freq OR PAF TGFb BRCA2 XRCC3 ERa 14% 7% 15% 20% 1.25 1.31 1.34 1.27 2.9% 2.1% 4.4% 5% 6.8% 7.4% 7.4% 6.8% 0.2% 0.3% 0.5% 0.5% Chk2 0.5% 2.4 0.6% 16% 0.5% ~2.0% BRCA2 N372H association with breast cancer risk Finns HH HDB HH UK set 3 HH UK set 2 HH UK set 1 HH Joint HH Joint NH Joint NN 0.1 1 10 p=0.02 Tee et al. In prep. 3133 Fiegelson et al. 2001 OR breast cancer Haiman et al. 1999 1081 Mitrunen et al. 2000 744 Kristensen et al. 1999 CYP17 t -34 c Spurdle et al. 2000 (cc Vs. tt) Miyoshi et al. 2000 Kuligina et al. 2000 Hamajima et al. 2000 310 Huang et al. 1999 Conclusion: This SNP has no main effect on breast cancer risk! Helzlouler et al. 1998 230 Weston et al. 1998 Bergman-Jungestrom et al. 1999 226 Young et al. 1999 Weston et al. 1998 N 0.1 1 10 100 Ye & Parry, 2002 Mutagenesis 17:119-126 Why a p value of p = 0.01 is not persuasive True association False association Prior probability of result (snp causing 1% of FRR, 100,000 snps in genome) 1/1000 999/1000 Probability given result has p = 0.01 99/100 1/100 99/100,000 999/100,000 Assuming random choice of ‘candidate’ gene only ~ 10% results at p = 0.01 are true (~50%, at p = 0.001) Summary of results 96 snps, 47 genes ~2000 cases, 2000 controls p-value 0.001 p = 0.01/0.0004 for comparison of distributions 0.01 0.05 observed 0.10 chance 1.00 0 10 20 30 40 50 SNP 60 70 80 90 100 % of excess FRR explained 0.5 1 1.3 2 relative risk Some reasons why human association studies may be difficult Inappropriate genetic models eg rare/multiple alleles Regulatory vs coding polymorphisms Numbers : inadequate statistical power Genetic background effects; interactions weak ‘main effect’, high-order interactions ‘null’ result = balance of susceptible and resistant on different BG Phenotypic heterogeneity eg ER+/ER-; histology Cancer/no cancer endpoint lacks power Intermediate phenotypes Serum estradiol and CYP19 Exon 10 t>c 3’UTR 20 18 Serum SHBG and SHBG Exon 8 g>a or D356N 60 50 16 40 14 30 12 10 tt tc P homogeneity = 0.0005 P trend <0.0001 cc 20 gg ga aa P homogeneity = 0.006 P trend = 0.006 (Ponder, Dowsett labs; EPIC; unpublished) Implications for breast cancer risk 2 fold increase in estradiol 30% increase in risk of breast cancer tt genotype of CYP19 c>t associated with 14% increase in estradiol: equivalent to 1.04 fold increase in breast cancer risk Where next? Empirical vs candidate approaches Snp genotyping now ~17c/genotype : ? screen 600 “enriched” cases/600 controls vs 1150 coding snps ~$240,000 Candidate gene approaches Candidates from cell biology Epidemiology Regulatory variants Quantitative phenotypes Leads from mouse models Mouse/human collaborations 1. Candidate susceptibility genes/regions mapped in susceptible/resistant crosses refined by amplicons/deletions in tumours allele-specific differences in expression/somatic change (easier in mouse because extended haplotypes) loci involved in control of gene regulation loci influencing intermediate phenotypes set up large cross and score multiple phenotypes How tightly should the region be defined? 300 kb Say 5 genes First pass = find all coding region snps at >5% Construct haplotypes, select minimum snp set = ? 30 snps Genotype 30 snps in 2000 cases/2000 controls = 120,000 genotypes Genotyping cost ~$20,000 @ 17c/genotype BUT : currently requires ~1000 snps at a time Mouse/human collaborations 2. Interactions Identification of interacting loci potentially approachable in mouse Develop and evaluate programmes to search for higher order interactions; ? applicability to man Mouse/human collaborations 3. Stages of cancer development ? Distinguish loci that influence multiplicity latency; progression invasion metastasis and resistance to these ? Loci that affect treatment response Mouse/human collaborations 4. “End game” - which is the active gene, snp? strain comparisons of variants dissection of complex QTLs transgenic models A new horizon in medicine? “‘Risk factor’ analysis will facilitate environmental modification, screening and therapeutic management of people before they develop symptoms” (Bell, BMJ 1998) “Differences in social structure, lifestyle and environment account for much larger proportions of disease than genetic differences …… Those who make medical and scientific policies ….. would do well to see beyond the hype” (Holtzman & Marteau, NEJM 2000) Strangeways Research Laboratories University of Cambridge Bruce Ponder Paul Pharoah Alison Dunning Fabienne Lesueur Bettina Kuschel Annika Auranen Katie Healey Craig Luccarini Jenny He Louise Tee Gary Dew Doug Easton Antonis Antoniou Mitul Shah Julian Lipscombe Nick Day; EPIC UCSF Allan Balmain Mandy Toland Joe Gray Mark Sternlicht NCI Kent Hunter Biochemistry, Cambridge Jim Metcalfe Cancer Research UK; MRC TGFb t/c -509 t 0.25 Pro/Leu 10 P tt vs cc c P c L 0.11 0.60 PP vs LL OR 1.25 (1.1 - 1.4) p = 0.01 OR 1.30 (1.1 - 1.5) p = 0.01 Which SNP is the functional variant? tt ProPro Pro10 homozygotes have increased risk regardless of c-509t genotype ct ProPro cc ProPro ct LeuPro cc LeuPro cc LeuLeu 0.1 1.0 Odds Ratio 10 TGFb in vitro secretion Time Course End Point 4 Pro10 3 Ratio P:L TGFb1 ng/ml 2 Leu10 1 0 0 6 12 18 hours (Metcalfe, Ponder labs, 2002) Funnel Plot For TGFb L10P O R (PP Vs. LL) N * Cohort study 4517 ABC 875 HDB 939 Finn 404 Hishido et al. * 3075 Ziv et al. 238 Frei 146 cases 2929 controls 0.1 1 10