* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Class Slides
Nutriepigenomics wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Gene expression profiling wikipedia , lookup
Genome (book) wikipedia , lookup
Point mutation wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Public health genomics wikipedia , lookup
Neuronal ceroid lipofuscinosis wikipedia , lookup
Protein moonlighting wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
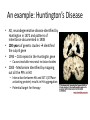
Protein Interactions and Disease Audry Kang 7/15/2013 Central Dogma of Molecular Biology Protein Review • Primary Structure: Chain of amino acids • Secondary Structures: Hydrogen bonds resulting in alpha helix, beta sheet and turns • Tertiary Structure: Overall Shape of a single protein molecule • Quaternary Structure: structure formed by several protein subunits What is “Protein Interaction?” • Physical contact between proteins and their interacting partners (DNA, RNA) – Dimers, multi-protein complexes, long chains – Identical or heterogeneous – Transient or permanent • Functional Metabolic or Genetic Correlations – Proteins in the same pathway or cycles or cellular compartments Protein-protein interactions • Nodes represent proteins • Lines connecting then represent interactions between them • Allows us to visualize the evolution of proteins and the different functional systems they are involved in • Allows us to compare evolutionarily between species Figure 1. A PPI network of the proteins encoded by radiation-sensitive genes in mouse, rat, and human, reproduced from [89]. Why Do We Care about PPI? • Proteins play an central role in biological function • Diseases are caused by mutations that change structure of proteins • Considering a protein’s network at all different functional levels (pair-wise, complexes, pathways, whole genomes) has advanced the way that we study human disease An example: Huntington’s Disease • AD, neurodegenerative disease identified by Huntington in 1872 and patterns of inheritance documented in 1908 • 100 years of genetic studies identified the culprit gene • 1993 – CAG repeat in the Huntingtin gene – Causes insoluble neuronal inclusion bodies • 2004 - Mechanism Identified by mapping out all the PPIs in HD – Interaction between Htt and GIT1 (GTPaseactivating protein) results in Htt aggregation – Potential target for therapy Experimental Identification of PPIs: Biophysical Methods – Provides structural information – Methods include: X-ray crystallography, NMR spectroscopy, fluorescence, atomic force microscopy – Time and resource consuming – Can only study a few complexes at a time Experimental Identification of PPIs: High-Throughput Methods Direct high-throughput methods: Yeast two-hybrid (Y2H) -Tests the interaction of two proteins by fusing a transcription-binding domain -If they interact, the transcription complex is activated -A reporter gene is transcribed and the product can be detected Drawbacks: -Can only identify pair-wise interactions -Bias for unspecific interactions http://www.specmetcrime.com/noncovalent_complexes_in_mass_s.htm Experimental Identification of PPIs: High-Throughput Methods Indirect high-throughput methods: • Looks at characteristics of genes encoding interacting partners • Gene co-expression – genes of interacting proteins must be co-expressed – Measures the correlation coefficient of relative expression levels • Synthetic lethality – introduces mutations on two separate genes which are viable alone but lethal when combined Drawbacks of Experimental Identification Methods • High false positive • Low agreement when studied with different techniques • Only generates pair-wise interaction relationships and has incomplete coverage Computational Predictions of PPIs • • • • Fast, inexpensive Used to validate experimental data and select targets for screening Allows us to study proteins in different levels (dimer, complex, pathway, cells, etc) Two categories: – Methods predicting protein domain interactions from existing empirical data about protein-protein interactions • • • Maximum likelihood estimation of domain interaction probability Co-expression Network properties – Methods relying on theoretical information to predict interactions • • • • Mirrortree Phylogenetic profiling Gene neighbors methods The Rosetta Stone Method Example: Theoretical Predictions of PPIs Based on Coevolution at the Full-Sequence Level The Principle: • Changes in one protein result in changes in its interacting partner to preserve the interaction • Interacting proteins coevolve similarly The Mirrortree Method • • • Measures coevolution for a pair of proteins Mirrortree correlation coefficient is used to measure tree similarity Each square is the tree distance between two orthologs (darker colors represent closeness) Method: 1. Identifies orthologs of proteins in common species 2. Creates a multiple sequence alignment (MSA) of each protein and its orthologs 3. Builds distance matrices 4. Calculated the correlation coefficient between distance matricies Protein Networks and Disease Studying the Genetic Basis of Disease • The correlation between mutations in a person’s genome and symptoms is not clear… • Pleiotrophy – single gene produces multiple phenotypes mutations in a single gene may cause multiple syndromes or only affects certain processes • Genes can influence one another – Epistasis – interact synergistcally – Modify each other’s expression • Environmental factors Studying the Molecular Basis of Disease • Crucial for understanding the pathogenesis and disease progression of disease and identifying therapeutic targets Role of protein interactions in disease • Protein-DNA Interaction disruptions (p53 TSP) • Protein Misfolding • New undesired protein interactions (HD, AD) • Pathogen-host protein interactions (HPV) Using PPI Networks to Understand Disease • PPI Networks can help identify novel pathways to gain basic knowledge of disease • Explore differences between healthy and disease states • Prediction of genotype-phenotype associations • Development of new diagnostic tools for identifying genotype-phenotype associations • Identifying pathways that are activated in disease states and markers for prognostic tools • Development of drugs and therapeutic targets