* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download BPM§1.2_Protein Struktur.key
Rosetta@home wikipedia , lookup
Protein design wikipedia , lookup
Bimolecular fluorescence complementation wikipedia , lookup
Protein purification wikipedia , lookup
List of types of proteins wikipedia , lookup
Protein mass spectrometry wikipedia , lookup
Western blot wikipedia , lookup
Protein folding wikipedia , lookup
Protein moonlighting wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Trimeric autotransporter adhesin wikipedia , lookup
Structural alignment wikipedia , lookup
Homology modeling wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Circular dichroism wikipedia , lookup
Protein domain wikipedia , lookup
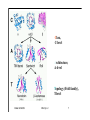
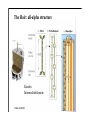
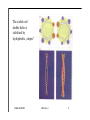
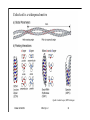
Hierarchy of protein structure Branden & Tooze, Introduction to Protein Structure Rädler WS2009 BPM §1.1.2 1 Secondary structure elements “Buchstaben” α-helix helix bundle β-strand β -sheet Rädler WS2009 BPM §1.1.2 2 Main supersecondary structure elements Rädler WS2009 BPM §1.1.2 “Worte” 3 Evolution from fragments - solenoid proteins “Worte” Quelle: Andrei Lupas, MPI Tuebingen Rädler WS2009 BPM §1.1.2 4 Rubredoxin Prealbumin (greek key) Phosphate Isomerase Rädler WS2009 BPM §1.1.2 5 Fold classification (protein taxonomy) classification: clustering proteins into structural families http://www.cathdb.info/latest/index.html Class, C-level Three major classes are recognised; mainly-alpha, mainly-beta and alpha-beta. Architecture, A-level This describes the overall shape of the domain structure as determined by the orientations of the secondary structures but ignores the connectivity between the secondary structures. Topology (Fold family), T-level Structures are grouped into fold families at this level depending on both the overall shape and connectivity of the secondary structures. Homologous Superfamily, H-level Rädler WS2009 This level groups together protein domains which are thought to share a common ancestor. BPM §1.1.2 6 Class, C-level Architecture, A-level Topology (Fold family), T-level Rädler WS2009 BPM §1.1.2 7 The Hair: all-alpha structure Keratin Intermediärfilament Rädler WS2009 BPM §1.1.2 8 The coiled-coil double helix is stabilized by hydrophobic „stripes“ Rädler WS2009 BPM §1.1.2 9 Coiled-coil is a widespread motive Quelle: Andrei Lupas, MPI Tuebingen Rädler WS2009 BPM §1.1.2 10 Silk : all beta structure Rädler WS2009 BPM §1.1.2 11 Antecedent domain segments encoded by mini-genes Extinct intermediates Present-day proteins N N N DsbA N N N Hetero-dimer N Fused dimer N N Thioredoxin N Gottschalk/SS 2005 BPMPonting §1.1.2 and Russell, J.Struct.Biol. 134:191-203 (2001) Lupas, Divergent evolution: Circular permutation IV Circular permutation in four easy steps: Tandem, in-frame gene duplication: Loss of stop codon: 3' deletion: Resolution through further deletion I: Gene returns to previous state II: Circular permutation Quelle: Andrei Lupas, MPI Tuebingen BPM §1.1.2 Divergent evolution: Deletions Grishin N.V., J.Struct.Biol. 134:167-185 (2001) BPM §1.1.2 Divergent evolution: Strand invasion I Grishin N.V., J.Struct.Biol. 134:167-185 (2001) BPM §1.1.2 Divergent evolution: Swaps I Grishin N.V., J.Struct.Biol. 134:167-185 (2001) BPM §1.1.2 Divergent evolution: Circular permutation I Grishin N.V., J.Struct.Biol. 134:167-185 (2001) BPM §1.1.2 Divergent evolution: Circular permutation II Grishin N.V., J.Struct.Biol. 134:167-185 (2001) BPM §1.1.2 TolC Rädler WS2009 BPM §1.1.2 19Tuebingen Quelle: Andrei Lupas, MPI