* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download NMR experiment-driven modeling of biological macromolecules
List of types of proteins wikipedia , lookup
Gene expression wikipedia , lookup
Protein (nutrient) wikipedia , lookup
Multi-state modeling of biomolecules wikipedia , lookup
Bottromycin wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Protein moonlighting wikipedia , lookup
Western blot wikipedia , lookup
Protein adsorption wikipedia , lookup
Interactome wikipedia , lookup
Proteolysis wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Drug design wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Pharmacometabolomics wikipedia , lookup
Protein structure prediction wikipedia , lookup
Homology modeling wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
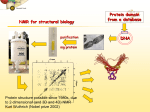
NMR experiment-driven modeling of biological macromolecules Torsten Herrmann Institut des Sciences Analytiques, Centre de RMN à très Hauts Champs, Université de Lyon/ UMR 5280 CNRS / ENS Lyon / UCB Lyon 1, France Nuclear Magnetic Resonance Spectroscopy (NMR) is one of the more versatile experimental techniques that allow determining three-dimensional (3D) structures of biomacromolecules at atomic resolution, whether these are proteins, RNA, DNA, and their complexes. Knowledge of the 3D structure is vital for understanding functions and mechanisms of action of macromolecules, and for rationalizing the effect of mutations. 3D structures are also important as guides for the design of new experimental studies and as starting point for rational drug design. Little more than ten years ago, protein NMR structure determination projects were framed in terms of months if not years of laborious, interactive work. Nowadays, owing to stunning advances in NMR experiments, instrumentation and notably computational data analysis, a relatively propitious protein candidate may be solved in a few weeks. Not surprisingly, around half of all protein structures solved in the Protein Data Bank using NMR have been thanks to automated, computational approaches. Here we will discuss emerging methods at the intersection of experimental NMR and computational analysis and prediction. Special emphasis will be placed on the development of computational approaches that are able to handle large amount of experimental data efficiently and that integrate with data from other experimental techniques. The latter aspect is essential for applications of NMR to advanced and challenging problems in systems biology.



![report [7]](http://s1.studyres.com/store/data/022227245_1-12cd315519aff78edcf9ef780e94c39c-150x150.png)