* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Histone modifications
Survey
Document related concepts
Transcript
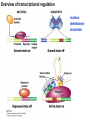
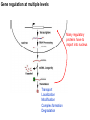
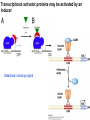
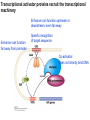
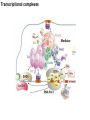
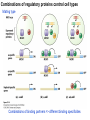
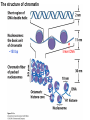
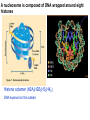
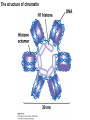
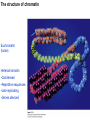
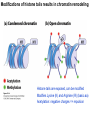
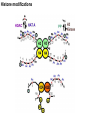
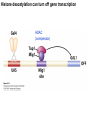
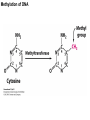
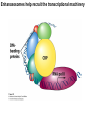
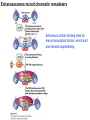
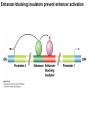
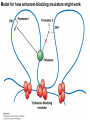
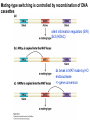
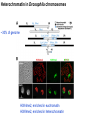
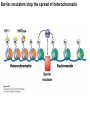
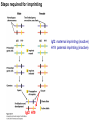
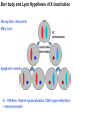
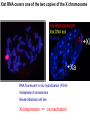
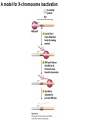
Griffiths • Wessler • Carroll • Doebley Introduction to Genetic Analysis TENTH EDITION CHAPTER 12 Regulation of Gene Expression in Eukaryotes © 2012 W. H. Freeman and Company CHAPTER OUTLINE 12.1 12.2 12.3 12.4 12.5 Transcriptional regulation in eukaryotes: an overview Lessons from yeast: the GAL system Dynamic chromatin Short-term activation of genes in a chromatin environment Long-term inactivation of genes in a chromatin environment 12.6 Gender-specific silencing of genes and whole chromosomes 12.7 Post-transcriptional gene repression by miRNAs The first cloned mammal Dolly and Bonnie Dolly, the Finn Dorset lamb in 1996 and her surrogate Scottish Blackface mother Ian Wilmut Dolly in Royal Museum of Scotland Dolly Dolly Parton Cells differ in gene expression All cells have the same genome, but each cell expresses only a subset of all genes globin myosin housekeeping erythrocyte muscle cell All genes Overview of transcriptional regulation nucleus (membrane) chromatin Gene regulation at multiple levels Many regulatory proteins have to import into nucleus Transport Localization Modification Complex formation Degradation Promoter-proximal elements precede the promoter of a eukaryotic gene Promoter-proximal elements are necessary for efficient transcription Point mutations throughout the promoter region were analyzed for their effects on transcription rates. The height of each line represents the transcription level relative to a wild-type promoter or promoter-proximal element (1.0). Transcription factors need multiple functional domains 1. A domain that recognizes a DNA regulatory sequence (the protein’s DNA-binding site) 2. A domain that interacts with one or more proteins of the transcriptional apparatus (RNA polymerase or a protein associated with RNA polymerase) 3. A domain that interacts with proteins bound to nearby regulatory sequences on DNA such that they can act cooperatively to regulate transcription 4. A domain that influences chromatin condensation either directly or indirectly 5. A domain that acts as a sensor of physiological conditions within the cell Direct or indirect (by interacting with other proteins) Model Organism Yeast Saccharomyces cerevisiae Budding yeast Brewer’s yeast Baker’s yeast The Gal pathway Expressed at low level Induced by galactose Regulated by Gal4 Transcriptional activator proteins bind to UAS elements in yeast UAS: Upstream Activation Sequences Binding site for Gal4 Can be far from promoter Transcriptional activator proteins are modular Reporter gene domain swap Transcriptional activator proteins may be activated by an inducer Galactose: inducing signal Transcriptional activator proteins recruit the transcriptional machinery Enhancer can function upstream or downstream, even far away Enhancer can function far away from promoter Specific recognition of target sequence Co-activator: Does not directly bind DNA Transcriptional complexes Combinations of regulatory proteins control cell types Mating type Combinations of binding partners => different binding specificities The structure of chromatin ~150 bp linker DNA A nucleosome is composed of DNA wrapped around eight histones Histone octamer (H2A2H2B2H32H42) DNA exposed on the outside The structure of chromatin The structure of chromatin Euchromatin (loose) Heterochromatin •Condensed •Repetitive sequences •Late replicating •Genes silenced Chromatin remodeling exposes regulatory sequences Linker DNA: sensitive to nuclease Nucleosomal DNA: protected from nuclease digestion Use nuclease sensitivity to determine chromatin state (open/closed) or nucleosome position + SWI-SNF + ATP Shifting of nucleosome position Exposes regulatory sequences The SWI-SNF complex for chromatin remodeling Yeast mutant screen sugar nonfermenting (snf) Mating type switch (swi) swi2=snf2 swi2/snf2 (“switch-sniff”) locus SWI-SNF complex Modifications of histone tails results in chromatin remodeling Histone tails are exposed, can be modified Modifies Lysine (K) and Arginine (R) (basic aa) Acetylation: negative charges => repulsion Histone modifications Histone modifications Acetylation of histones Histone acetyltransferase (HAT) Histone de-acetylase (HDAC) Histone modifications Histone code Alternative modifications on the same residue Regulation of gene expression by histone acetylation Histone deacetylation can turn off gene transcription HDAC (corepressor) Inheritance of chromatin states Epigenetic memory: heritable traits (over rounds of cell division and sometimes transgenerationally) that do not involve changes to the underlying DNA sequence. (e.g. chromatin state) Methylation of DNA A model for the inheritance of DNA methylation In mammals, 70-80% of CG are methylated genome-wide. CpG island: clusters around gene promoter maintenance hemi-methylated Enhanceosomes help recruit the transcriptional machinery Enhanceosomes recruit chromatin remodelers Enhancers contain binding sites for many transcription factors, which bind and interact cooperatively. Enhancer-blocking insulators prevent enhancer activation Model for how enhancer-blocking insulators might work Mating-type switching is controlled by recombination of DNA cassettes silent information regulators (SIR) Sir2 (HDAC) ds break in MAT made by HO endonuclease => gene conversion Gene silencing is caused by the spread of heterochromatin Position-effect variegation (PEV) w+ is expressed in some cells => not a mutation in w gene Clonal => epigenetic memory Heterochromatin in Drosophila chromosomes ~30% of genome H3K4me2, enriched in euchromatin H3K9me2, enriched in heterochromatin Some genes enhance or suppress the spread of heterochromatin Enhancer Suppresor Su(var)2-5 = HP1 (heterochromatic protein 1) Su(var)3-9 = histone methylatransferase Multiple states of Lysine methylation Heterochromatin may spread farther in some cells than in others Barrier insulators stop the spread of heterochromatin Genomic imprinting Genomic imprinting Phenotype depends on the parental origin of the genes Inactivation of genes and chrosomomes Genomic imprinting mouse/human ~100 imprinted genes No changes in DNA sequence Genomic imprinting requires insulators imprinting control region DNA methylation Unusual inheritance of imprinted genes Steps required for imprinting Igf2: maternal imprinting (inactive) H19: paternal imprinting (inactive) Igf2 H19 X inactivation Dosage compensation for X chromosome female: XX male: XY Barr body and Lyon Hypothesis of X inactivation Murray Barr: discoverer Mary Lyon Epigenetic memory Xi: H3K9me, histone hypoacetylation, DNA hypermethylation ~ heterochromatin Xist non-coding Xist RNA covers one of the two copies of the X chromosome RNA fluorescent in situ hybridization (FISH) metaphase chromosomes female fibroblast cell line Xist expression => cis-inactivation A model for X-chromosome inactivation Possible models for the repression of translation by miRNA