* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Phenotypic Variance
Dual inheritance theory wikipedia , lookup
Genetically modified food wikipedia , lookup
Fetal origins hypothesis wikipedia , lookup
Medical genetics wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Gene expression programming wikipedia , lookup
Group selection wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Public health genomics wikipedia , lookup
Dominance (genetics) wikipedia , lookup
Polymorphism (biology) wikipedia , lookup
Genetic drift wikipedia , lookup
Genetic engineering wikipedia , lookup
Genome (book) wikipedia , lookup
Koinophilia wikipedia , lookup
History of genetic engineering wikipedia , lookup
Designer baby wikipedia , lookup
Human genetic variation wikipedia , lookup
Population genetics wikipedia , lookup
Behavioural genetics wikipedia , lookup
Microevolution wikipedia , lookup
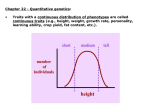
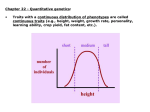
Quantitative Genetics and Multifactorial Traits A field of pumpkins, where size is under the influence of additive alleles I. Quantitative Characteristics Vary Continuously and Many Are Influenced by Alleles at Multiple Loci Qualitative characteristics Quantitative characteristics 2 Quantitative (polygenic) traits whose phenotypes result from both gene action and environmental influences are often termed multifactorial or complex traits 3 Not All Polygenic Traits Show Continuous Variation Meristic traits: the phenotypes are described by whole numbers The number of seeds in a pod, eggs laid by a chicken Threshold traits: present or absent 4 The Multiple Gene Hypothesis for Quantitative Inheritance P1 F1 Nilsson-Ehle , 1909 Kernel Color in Wheat The results of crossing individuals heterozygous for different numbers of loci affecting a characteristic 6 Determining Gene Number for a Polygenic Characteristic Assumption: • All the alleles contribute equally and additively; • Phenotypes of F2 is not affected significantly by environments. 7 II. Statistical Methods Are Required for Analyzing Quantitative Characteristics 8 Relations among the methods of studying quantitative genetics. 9 Distribution Phenotypic variation in a group can be conveniently represented by a frequency distribution Numbers or proportions of individuals phenotypic classes 10 Distributions of phenotypes can assume several different shapes Normal distribution Skewed (不对称) and bimodal (双峰) 11 Samples and Populations Samples Be representative of the whole population Be large enough 12 The Mean The mean (average) provides information about the center of the distribution 13 The Variance and Standard Deviation Variance (方差) The variability of a group of measurements The variance (s2) is defined as the average squared deviation from the mean. 14 Standard deviation (s) (标准差) The standard deviation is often preferred for describing the variability of a measurement. The proportions of a normal distribution occupied by plus or minus one, two, and three standard deviations from the mean 15 Analysis of a quantitative character • One of the tomato homozygous varieties produces fruit averaging 18 oz. in weight, whereas fruit from the other averages 6 oz. • Crossed these two homozygous varieties. F1 Distribution of tomato weight F2 626/52=12.04 872/72=12.11 1.29 4.27 1.13 2.06 12.04±1.13 12.11±2.06 The number of genes that control fruit: F2: 1/4n=1/72, n=3~4 Correlation The relation between two characteristics is called a correlation Correlations between characteristics are measured by a correlation coefficient (r ) Covariance (cov) (协方差) 17 The correlation coefficient describes the relation between two or more variables 18 19 Regression (回归) The regression line can be represented by a is the y intercept of the line, which is the expected value of y when x is 0. b is the slope of the regression line, also called the regression coefficient (回归系数) 20 The regression equation (y = a + bx ) can be used to predict the value of any y given the value of x. 21 III. Heritability Is Used to Estimate the Proportion of Variation in a Trait That Is Genetic The proportion of the total phenotypic variation that is due to genetic differences is known as the heritability 22 Phenotypic Variance (VP) Components of phenotypic variance Genetic variance: VG Environmental variance: VE Genetic−environmental interaction variance: VGE 23 Components of genetic variance Additive genetic variance: V A Dominance genetic variance:V D Genic interaction variance:V I 24 Types of Heritability (遗传率) Broad-sense heritability (H 2): Narrow-sense heritability (h 2): 25 Calculating Heritability Heritability by elimination of variance components In theory, we can make V E = 0 by raising all individuals in exactly the same environment, then VGE=0, VP=VG Instead, we can make V G = 0 by raising genetically identical individuals, then VGE=0, VP=VE 26 Ex am ple: Estimate the heritability of white spotting in guinea pigs Measured the phenotypic variance for white spotting in a genetically variable population and found that VP= 573 Inbred the guinea pigs for many generations to get homozygous and genetically identical individuals. measured their phenotypic variance and got VP=340=VE. 27 Heritability by parent–offspring regression Heritability and degrees of relatedness ……. 28 Artificial Selection Artificial selection is the process of choosing specific individuals with preferred phenotypes from an initially heterogeneous population for future breeding purposes. Alleles that generate dominance effects (V D ) or interact epistatically (V I ) are less responsive to artificial selection. Narrow-sense heritability,h 2 , can be used to predict the impact of selection 29 How to calculate narrow-sense heritability h 2 ? The simplest approach is to select individuals with superior phenotypes for the desired quantitative trait from a heterogeneous population and breed offspring from those individuals. selection response (R) selection differential (S) M : mean score for the trait of the original population M1: mean score of the selected individuals used as parents M2: mean score of those offspring Realized heritability 30 孵化率 受孕率 31 Theoretically, the process will continue until all individuals in the population possess a uniform genotype that includes all the additive alleles responsible for high oil content. At that point, h 2 →0 Response of corn selected for high and low oil content over 76 generations. The numbers in parentheses at generations 9, 25, 52, and 76 for the "high oil line” indicate the calculation of heritability at these points in the continuing experiment. 32 Artificial selection has produced the tremendous diversity of shape, size, color, and behavior seen today among breeds of domestic dogs. 33 Prediction Phenotypes Ex am ple : IQ score prediction • Mother: 110 • Father: 120 • Mean of population: 100 IQs of their offsprings? Narrow-sense heritability (h2) of IQ=0.4 The predicted IQ value will be: 106 if h2 =0.4 115 if h2 =1 34 The Limitations of Heritability Heritability does not indicate the degree to which a characteristic is genetically determined An individual does not have heritability There is no universal heritability for a characteristic Even when heritability is high, environmental factors may influence a characteristic Heritabilities indicate nothing about the nature of population differences in a characteristic 35 IV. Quantitative Trait Loci (QTLs) A chromosome region is identified as containing one or more genes contributing to a quantitative trait is known as a quantitative trait locus (QTL) 36 A generic case of QTL mapping • Individuals from highly divergent lines created by artificial selection are chosen as parents • These individuals are crossed to produce F1 and F2 generations. F2 generations carry different portions of the P1 genome with different QTL genotypes and associated phenotypes. The F2 is known as the QTL mapping population. • Measure phenotype of the trait among individuals and identify genomic differences among individuals by using DNA markers. • Computer-based statistical analysis is used to search for linkage between the markers and a phenotypic variation. • If a DNA marker is linked to a QTL, the marker locus and the QTL are cosegregate. • When numerous QTLs for a given trait have been located, a genetic map is created. 37 Annual Review of Genetics, Vol . 27:205-233,1993 38 Methods to identify QTLs for fruit weight in tomatoes Ancestral and modern-day tomato 39 RFLP and QTLs for fruit weight on four chromosomes in the tomato genome 40 • QTL mapping has identified more than 28 QTLs related to this variation in fruit weight • One QTL, called fw2.2 (on chromosome 2), was identified accounting for about 30% of the variation in fruit weight • A specific gene, ORFX, was identified within this QTL. A cloned ORFX gene from small-fruited varieties is transferred to a normally large tomatoes, the transgenic plant produces fruits with reduced weight. • ORFX gene encodes a protein that negatively regulates cell division during fruit development • Yet ORFX and other related genes cannot account for all the observed variation in tomato size -: control; +: transgenic 41