* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Quantitative Genetics and Multifactorial Traits
Heritability of autism wikipedia , lookup
Designer baby wikipedia , lookup
Pharmacogenomics wikipedia , lookup
Group selection wikipedia , lookup
Genetic engineering wikipedia , lookup
Genetic testing wikipedia , lookup
History of genetic engineering wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Polymorphism (biology) wikipedia , lookup
Genome (book) wikipedia , lookup
Genetic drift wikipedia , lookup
Koinophilia wikipedia , lookup
Public health genomics wikipedia , lookup
Medical genetics wikipedia , lookup
Human genetic variation wikipedia , lookup
Microevolution wikipedia , lookup
Population genetics wikipedia , lookup
Behavioural genetics wikipedia , lookup
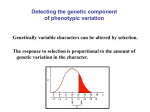
Quantitative Genetics and Multifactorial Traits The genetic analysis of complex characteristics is quantitative genetics Complex characteristics usually are either polygenic or are influenced by environmental factors Characteristics that are both polygenic and influenced by environmental factors are multifactorial Sometimes the relationship between genotype and phenotype is fairly easy to understand o In some instances, it is essentially additive o In the case of quantitative characteristics, the phenotypes are not distinct and can be quite hard to interpret But not all quantitative characteristics exhibit continuous phenotypic variation - simply because they can’t o Litter size has to yield whole numbers but the underlying genetics can exhibit quantitative influences (polygenic and environmental factors) o These are referred to as meristic characteristics Threshold characteristics can also be confusing o There are only two phenotypes exhibited but the underlying genetics exhibit quantitative influences o In this case, susceptibility is determined by a number of factors o When the susceptibility (liability or risk) is larger than the threshold level, the disease is seen Inheritance for a polygenic characteristic can be broken up into the inheritance for each individual gene The trick is that after you break the crosses apart, you need to put the results back together As the number of loci affecting a phenotype increases, the number of phenotypic classes increases even more Under some circumstances, we can estimate the number of genes involved o If we can begin with two parents who are homozygous for different alleles at each locus, the F1 will all be heterozygous o If we mate two F1, then (1/4)n will be the number of F2 individuals that should resemble each of the original homozygous parents Statistical Analysis We can use some basic statistical tools to analyze the genetics behind quantitative characteristics There are three common types of distribution curves Samples and populations o When sampling is completely random and large, it can provide information about the entire population Variance provides information on how spread out a distribution is o The greater the variance, the more spread out the distribution is Standard deviation o We can expect various percentages of the samples to fall within the ranges determined by the standard deviation Correlation refers to a relationship between two related characteristics o Correlation coefficient (r) o Covariance Correlation coefficients can range from -1 to +1 Coefficients of -1 or +1 indicate perfect correlations Coefficients near these values indicate strong correlations Coefficients near 0 indicate weak or no correlation Heritability o The proportion of phenotypic variation due strictly to genetics o Phenotypic variance o VP = VGenetic + VEnvironmental + VGE o Genetic variance can be further subdivided o VG = VA + VD + VI o VA = additive genetic variance o VD = dominance genetic variance o VI = genic interaction variance o Therefore, VP = (VA + VD + VI)+ VE + VGE o Broad-sense heritability (H2) o Can range from 0-1 o 0 = no genetic influence, all environmental o 1 = total genetic influence, no environmental o We can also consider how much is due to additive genetic variance o Narrow-sense heritability (h2) o This is especially useful to breeders because it determines how much the offspring will resemble the parents o Calculating heritability o Elimination of variance components Since VP = VG + VE + VGE, if we set VG=0, then VP = VE In practice, we might run two sets of experiments 1. One with genetically-identical individuals - This would give us VE (gen identical) 2. One with genetically-variable individuals - This would give us VP (gen variable) o VG (gen variable) = VP (gen variable) - VE (gen identical) For example, if VP (gen variable) = 573, and VP (gen identical) = 340 when VG (gen identical) = 0 o Then VP (gen identical) = VE (gen identical) o VP - VE = VG so 573 - 340 = 233 Heritability tells us how much of the phenotypic variance is due to genetic variance o Indicates the degree to which genes determine variation of a characteristic in the test group o It does not tell us the degree to which a characteristic is genetically determined o Heritability deals only with groups of individuals, not the individuals by themselves o The particular group is also important o Since heritability is unique for a particular population, it cannot be used to determine the reason for phenotypic differences between populations Response to selection o Artificial selection has been practiced by humans for centuries o The amount a characteristic subjected to selective pressure changes in one generation o response to selection = mean phenotype of the offspring - mean phenotype of the original population o Depends on 1. Narrow-sense heritability How much do the offspring resemble the parents? 2. Selection differential o Difference between the mean phenotype of the selected parents and the mean phenotype of the original population= R = h2 x S o R = response to selection; h2 = narrow-sense heritability; S = selection differential o R predicts the expected change in the offspring relative to the original population o Heritability determined by a response-to-selection experiment is termed the realized heritability o Eventually the response to selection will level off o We can also use individuals with different degrees of relatedness o Can compare results for monozygotic (MZ) twins vs. dizygotic (DZ) o H2 = 2(rMZ - rDZ) o Chromosomal regions that contain genes influencing quantitative traits are called quantitative trait loci (QTLs) o Mapping QTLs o This is a complicated task with many different factors to consider o Correlated responses o Phenotypic correlation is seen when two phenotypes are correlated o This can be due to environmental or genetic correlations o We can see examples of other correlated phenotypes in the domestication of dogs