* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download practice mid-term 1
Genetic code wikipedia , lookup
Oxidative phosphorylation wikipedia , lookup
Metabolic network modelling wikipedia , lookup
Point mutation wikipedia , lookup
Interactome wikipedia , lookup
Enzyme inhibitor wikipedia , lookup
Multi-state modeling of biomolecules wikipedia , lookup
Evolution of metal ions in biological systems wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Western blot wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Protein structure prediction wikipedia , lookup
Proteolysis wikipedia , lookup
Photosynthetic reaction centre wikipedia , lookup
Metalloprotein wikipedia , lookup
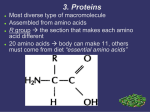
PRACTICE MID-TERM 1 01. Mark each statement below TRUE or FALSE and give reasons. You will get credit ONLY if your reasoning is correct. A) At equilibrium, there is no conversion of S P B) The rate of a reaction depends only on its G0 C) An enzyme’s binding energy determines the rate of the catalyzed reaction D) The energy of the E.S complex can sometimes be higher than the energy of the E.TS complex E) Increasing the temperature of a reaction will change the G0 02. Consider the two enzyme-catalyzed reactions below: Reaction 1: G0 for formation of the ES = -2 kJ mole-1 Reaction 2: G0 for formation of the ES = -4 kJ mole-1 Can you predict which of these reactions will have a higher rate? Explain your answer. 03. A reaction has a G0 = 10 kJ mol-1. At equilibrium, what is the ratio of [Products]:[Reactants]? 04. In a cell, Keq for a reaction is 344. Is this a spontaneous reaction in the cell? Give reasons for your answer. 05. A mutation in a protein converts a Cys Ala. What are all the possible effects this mutation might have on the protein’s structure? Mention the effects on secondary, tertiary and quaternary structure. Give reasons for your answer. 06. A mutation from Gly Phe has no effect on the activity of an enzyme. What are some reasons to account for this? 07. Which of the following statements is TRUE about secondary structure (mark all that apply)? A) B) Stabilized by hydrophobic interactions Without constraints on bond rotation angles, no ordered secondary structures would form C) A given protein cannot contain both parallel and anti-parallel -sheets D) Bends and loops are not important in forming secondary structures 08. Two proteins have a pI of 7.6. Therefore: A) They must have the same amino acid seqence B) They must have the same 3° structure C) They must have the same molecular mass D) They will have not move in an electric field at pH 7.6 PRACTICE MID-TERM 1 09. For each molecule shown, in the box below the molecule, write in the amino acid it depicts. If the molecule shown is not a standard amino acid, write in “NS.” 10. With regards to the Keq, the effect of an enzyme on a chemical reaction is BEST described as: A) Increasing the Keq in less time B) Decreasing the Keq in less time C) Maintaining the Keq for less time D) Achieving the Keq in less time 11. A schematic of an enzyme active site is shown. Based on this, which of the following will be the MOST LIKELY tripeptide substrate for this enzyme. Explain your answer. A) Ile-Glu-Ala B) Val-Leu-Lys C) Ile-Ile-Asp D) Glu-Glu-Tyr 12. Protein X by itself tends to form large aggregates in solution. However, addition of only a very small amount of another protein, Y causes Protein X to become soluble, without forming any aggregates. What is the MOST LIKELY explanation for these observations? A) Protein Y is a chaperone, which refolds Protein X, causing hydrophobic regions of X to be buried internally B) Protein Y causes the release of more water molecules from the solvation shell, increasing entropy of the solution C) Protein Y causes an increase in the number of hydrophilic residues of Protein X, increasing its solubility D) Protein Y adds amino acids to Protein X, changing its size and structure PRACTICE MID-TERM 1 13. A protein is made of an ɑ-helix, consisting almost exclusively of hydrophobic amino acids. Where in the cell are you MOST LIKELY to find this protein? Why? A) In the plasma membrane where the hydrophobic R-groups can interact with the lipids B) In the ER, where these is less water to interact with the hydrophobic R-groups of the protein C) In the lysosome where the acidic pH means the protein can fold differently because of different charges on the R-groups D) In the nucleus where the R-groups can form hydrophobic interactions with the nucleotides in DNA 14. Consider the reaction depicted in the graph. Based on this graph, can I conclude that this reaction MOST LIKELY does not have a transition state? Why or why not? Can I conclude that MOST LIKELY this reaction has reached equilibrium? Why or why not? (HINT: Note the labels on the axes) 15. Consider the uncatalyzed reaction A B. If I start with 1 mole of A, at equilibrium, I am left with 0.8 moles of A. It takes 30 minutes for this reaction to reach equilibrium. For the same reaction catalyzed by an enzyme: A) MOST LIKELY, what can you predict about how long the reaction will take to reach equilibrium? B) MOST LIKELY, how much of A will be left at the end of the reaction if I started with 1 mole of A? 16. Consider the reaction catalyzed by enzyme “E” shown in the graph. I take 1 mole of “R” and add the enzyme “E” to it. What is the MOST LIKELY outcome? A) The R Q reaction will reach equilibrium faster than the uncatalyzed reaction B) Nothing will happen since the R Q reaction is nonspontaneous C) The uncatalyzed reaction will proceed at the same rate as the catalyzed reaction since E does not catalyze the R Q reaction D) Nothing will happen since E does not catalyze the R Q reaction PRACTICE MID-TERM 1 17. I compare the sequence of Protein “G” from humans and gorillas. The differences are shown schematically in the figure (amino acid residues that are the same in both species are NOT labelled; amino acids present in the active site are labelled with a “*”). Based on this, MOST LIKELY what can you conclude about the function of Protein “G” in the two species? A) The amino acids changed are similar in their hydrophobicity and hydrophilicity; therefore, the proteins will function the same B) The amino acids in the active site are different; therefore, the functions will be different C) We cannot predict anything about the function, since we do not know the overall structure of the two proteins D) The active site residues have opposite charges; therefore, the overall function will be conserved, but the protein will interact with an oppositely charged substrate 18. Consider the molecule shown. It has a pK1 of 2.3 and a pK2 of 3.6. What is the overall charge on this molecule at pH 1? 19. Select the BEST choice regarding the statement below: Active sites of enzymes cannot have amino acids with hydrophobic R-groups. A) TRUE: hydrophbic residues will be on the inside of proteins, while active sites are on the outside B) TRUE: hydrophobic residues will prevent the substrate from binding the active site C) FALSE: weak interactions with hydrophobic residues can stabilize the transition state D) FALSE: hydrophobic residues cause water to be excluded from active sites, thereby allowing the substrate to bind 20. The measured activity of 1 mg of a protein in water is 455 units. The measured activity of 1 mg of the same protein in benzene (a non-polar solvent) is 2 units. What is the BEST explanation for this data? A) Non-polar solvents disrupt H-bonds required for secondary structure B) Non-polar solvents disrupt hydrophobic interactions required for tertiary structure C) Covalent peptide bonds are broken in non-polar solvents D) H-bonding and hydrophobic interactions are disrupted in non-polar solvents 21. Based on the data in the graph, MOST LIKELY, which line represents the catalyzed reaction? Why? A) Solid line is catalyzed reaction because it has lower activation energy B) Dotted line is catalyzed reaction because [Products] is higher C) Solid line is catalyzed reaction because it has a lower slope D) Dotted line is catalyzed reaction because it reaches equilibrium faster PRACTICE MID-TERM 1 22. Refer to the point marked “A” in the figure for the previous Question. What is your BEST conclusion about “A”? A) Reaction equilibrium; no change in [Products] B) Reaction equilibrium; no change in G C) Transition state; highest energy of reaction D) Transition state; extremely short-lived intermediate 23. Two enzymes that catalyze different reactions contain the same domain. Based on this observation, what is your BEST conclusion about these two enzymes? A) Extremely unlikely. Enzymes with similar domains should catalyze similar reactions B) The common domain is likely non-functional in these enzymes C) The two enzymes could have evolved from a common protein D) The common domain will most likely be mutated in one of the proteins 24. The figure depicts the primary structure of an enzyme. The amino acids that make up the active site are marked for the wild type enzyme (top amino acids) and a mutant enzyme (bottom amino acids). Based on this, what is your BEST prediction about the mutant enzyme? A) Mutant enzyme will have lower activity; Ile will not bind substrate as well as Leu, and will not stabilize the transition state B) Mutant enzyme will have lower activity; mutation will cause protein to unfold C) Mutant enzyme will have the same activity as wild type; Leu and Ile have similar Rgroups with similar weak bonds D) Mutant enzyme will have higher activity; Ile will accommodate the substrate better and stabilize the transition state better 25. The data in the graph was obtained by measuring the product formed over time with a wildtype and mutant enzyme. Based on these data, can you determine which line represents the wildtype and mutant enzyme? Why? A) Solid line is mutant enzyme; [Products] is lower B) Dotted line is mutant enzyme; reaches equilibrium faster C) Solid line is mutant enzyme; reaches equilibrium slower D) Cannot tell which is mutant or wild type; mutations can increase or decrease enzyme activity 26. Based on the data in graph, can you predict which reaction is spontaneous under these reaction conditions (1, 2 or both)? Why? A) Only Reaction 1 is spontaneous, Keq > 1 B) Both Reaction 1 and 2 are spontaneous, Keq is positive C) Only reaction 1 is non-spontaneous, G is positive D) Both reaction 1 and 2 are non-spontaneous, G is positive PRACTICE MID-TERM 1 27. A -> X + Y G = -29 kCal mol-1; Activation energy = 5 kCal mol-1 X -> D + E G = +3 kCal mol-1; Activation energy = 10 kCal mol-1 We can couple these two reactions to carry out the overall reaction: A -> Y + D + E. Which of the following statements is TRUE about these reactions? A) A -> X+Y will occur more slowly since its activation energy is lower B) The coupled reaction will be very slow since its activation energy will be 15 kCal mol -1 C) The coupled reaction will have a very low KM (<0.5) since its rate is very high D) X -> D+E is an endergonic reaction, that might be reversible 28. A protein is composed only of the amino acids Ala, Gly, Val, Leu, Ile, and Phe. This protein is insoluble in water, and forms large aggregates when placed in pure water. What is the BEST explanation for all this? 29. The G for the reaction D E is -23 kJ mol-1. I start off with 1M D and measure the [E] over time at constant temperature. The maximal solubility of D is 1M. My data is shown in the graph. MOST LIKELY, what did I do at the point marked “X” in the graph? Explain your answer. X