* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Blood 93/6 - Semantic Scholar
Organ-on-a-chip wikipedia , lookup
List of types of proteins wikipedia , lookup
Purinergic signalling wikipedia , lookup
Cell culture wikipedia , lookup
Tissue engineering wikipedia , lookup
Cell encapsulation wikipedia , lookup
Cellular differentiation wikipedia , lookup
Paracrine signalling wikipedia , lookup
VLDL receptor wikipedia , lookup
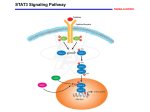
From www.bloodjournal.org by guest on April 18, 2017. For personal use only. HEMATOPOIESIS Cytoplasmic Domains of the Leukemia Inhibitory Factor Receptor Required for STAT3 Activation, Differentiation, and Growth Arrest of Myeloid Leukemic Cells By Mikio Tomida, Toshio Heike, and Takashi Yokota Leukemia inhibitory factor (LIF) induces growth arrest and macrophage differentiation of mouse myeloid leukemic cells through the functional LIF receptor (LIFR), which comprises a heterodimeric complex of the LIFR subunit and gp130. To identify the regions within the cytoplasmic domain of LIFR that generate the signals for growth arrest, macrophage differentiation, and STAT3 activation independently of gp130, we constructed chimeric receptors by linking the transmembrane and intracellular regions of mouse LIFR to the extracellular domains of the human granulocyte macrophage colonystimulating factor receptor (hGM-CSFR) ␣ and c chains. Using the full-length cytoplasmic domain and mutants with progressive C-terminal truncations or point mutations, we show that the two membrane-distal tyrosines with the YXXQ motif of LIFR are critical not only for STAT3 activation, but also for growth arrest and differentiation of WEHI-3B Dⴙ cells. A truncated STAT3, which acts in a dominant negative manner was introduced into WEHI-3B Dⴙ cells expressing GM-CSFR␣-LIFR and GM-CSFRc-LIFR. These cells were not induced to differentiate by hGM-CSF. The results indicate that STAT3 plays essential roles in the signals for growth arrest and differentiation mediated through LIFR. r 1999 by The American Society of Hematology. L Baumann et al8 constructed chimeric receptors consisting of the extracellular domain of the granulocyte colony-stimulating factor receptor (G-CSFR) and the cytoplasmic domain of LIFR and showed that the cytoplasmic domain of LIFR was capable of inducing gene expression in hepatic cells when induced to form a homodimer. It was not known whether or not the cell differentiation and proliferation signals were generated through the cytoplasmic region of LIFR alone. We fused the transmembrane and intracellular regions of LIFR to the extracellular parts of the human granulocyte-macrophage colony-stimulating factor receptor (GM-CSFR) ␣ and c chains, respectively. GMCSFR is composed of a specific GM-CSFR ␣ chain and a common c subunit that is shared with the receptors for IL-3 and IL-5.15 Using these chimeric receptors, we show that a homodimer of the cytoplasmic domain of LIFR can generate the signals for growth arrest and differentiation in mouse WEHI-3B D⫹ and M1 myeloid leukemic cells. Dimerization of the hematopoietin receptors initiates intracellular signaling by activating members of the receptor-associated tyrosine kinase family, referred to as Jaks.16 The information is next relayed by a family of transcription factors known as STATs (signal transducers and activators of transcription).17 Tyrosine phosphorylation of the STAT proteins is necessary for the formation of STAT homo- or heterodimers and subsequent DNA binding. IL-6 type cytokines preferentially activate STAT-3, STAT-1,18 and STAT-5.19 Tyrosine residues within receptors mediate signal transduction through the recruitment of molecules containing either Src homology 2 domains or phosphotyrosine binding domains.18 There are six tyrosine residues within the LIFR cytoplasmic domain.20,21 In this study, mutational analysis of LIFR showed the tight correlation between STAT3 activation and induction of differentiation of WEHI-3B D⫹ cells. Overexpression of dominant negative STAT3 blocks the induction of growth arrest and differentiation mediated through LIFR. EUKEMIA INHIBITORY factor (LIF)/differentiationstimulating factor (D-factor) is a multifunctional cytokine that was initially identified as a factor that inhibits the proliferation and induces macrophage differentiation of the murine myeloid leukemic cell line, M1.1-4 WEHI-3B D⫹ leukemic cells transfected with LIF receptor (LIFR) cDNA are also induced to differentiate by LIF.5 LIF maintains embryonic stem cells in an undifferentiated, pluripotent state, enhances the synthesis of acute phase proteins by hepatocytes, regulates nerve differentiation, and induces cardiac myocyte hypertrophy.6-11 It is a member of the interleukin (IL)-6 type cytokine family. Receptors for these cytokines are composed of multisubunit complexes that share a common signaling subunit, gp130. IL-6 and IL-11 induce the homodimerization of gp130, while LIF, cardiotrophin (CT)-1, ciliary neurotrophic factor (CNTF), and human oncostatinM(OSM) induce gp130 heterodimer formation with the LIFR subunit.12 The essential role of gp130 in the signaling by these receptors is demonstrated by the fact that antibodies to gp130 are capable of neutralizing the cell responses to all of these cytokines.12 Targeted disruption of the LIFR gene causes placental, skeletal, neural, and metabolic defects, and thereby results in perinatal death.13,14 However, the functional significance of the cytoplasmic domain of LIFR is not well understood. From the Saitama Cancer Center Research Institure, Ina, Saitama, Japan; and the Department of Stem Cell Regulation, Institute of Medical Science, University of Tokyo, Shirokanedai, Minato-ku, Tokyo, Japan. Submitted July 8, 1998; accepted November 5, 1998. Supported in part by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports and Culture of Japan. Address reprint requests to Mikio Tomida, PhD, Saitama Cancer Center Research Institute, Ina, Saitama 362-0806, Japan. The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked ‘‘advertisement’’ in accordance with 18 U.S.C. section 1734 solely to indicate this fact. r 1999 by The American Society of Hematology. 0006-4971/99/9306-0005$3.00/0 1934 MATERIALS AND METHODS Cells and cell culture. WEHI-3B D⫹ leukemic cells (kindly provided by Dr Alan C. Sartorelli, Yale University School of Medicine, New Haven, CT)) were cultured in McCoy’s 5A modified medium (GIBCO-BRL, Grand Island, NY) supplemented with 10% fetal calf serum (FCS). M1 cells were cultured as described previously.1,2 Blood, Vol 93, No 6 (March 15), 1999: pp 1934-1941 From www.bloodjournal.org by guest on April 18, 2017. For personal use only. ROLE OF STAT3 IN DIFFERENTIATION OF WEHI-3B Cytokines. Recombinant human GM-CSF was purchased from Peprotech, London, England. Recombinant human IL-6 was kindly provided by Ajinomoto Co (Kawasaki, Japan). Recombinant human D-factor (LIF) was produced in Chinese hamster ovary cells and purified to homogeneity as described previously.4 Plasmid construction. For the construction of pCAG-hGM-CSFR ␣-mLIFR and pMKIT-hGM-CSFR c-mLIFR, the cDNA of mouse LIFR was cloned from ES cell line A3.1 by reverse transcriptionpolymerase chain reaction (RT-PCR).21,39 The PCR products were inserted into pSP72 (Promega, Madison, WI) at the Pvu II site and the nucleotide sequence was confirmed using a 373A DNA sequencing system (Perkin Elmer, Foster City, CA). The hGM-CSFR␣ (pCEV4hGMR␣) and hGM-CSFRc (pME18S-KH97) cDNAs were kindly provided by Dr A. Miyajima (University of Tokyo, Tokyo, Japan). All of these cDNAs were subcloned into pCAG22 at the Xho I site, which was provided by Dr J. Miyazaki (Osaka University, Osaka, Japan). The chimeric receptor constructions, which have the extracellular domain of hGM-CSFR␣ or c linked to the transmembrane and cytoplasmic regions of mLIFR, were generated by overlap extension using PCR.23 In brief, complementary oligonucleotide primers and PCR were used to generate two DNA fragments with overlapping ends. In the case of hGM-CSFR␣-mLIFR, the following two components were first generated by PCR; the extracellular domain fragment of hGM-CSFR␣ with the first 9 bp of the transmembrane domain of mLIFR and the transmembrane/intracellular domain of mLIFR with the last 9 bp of the extracellular domain of hGM-CSFR␣. These fragments were combined in a subsequent fusion reaction, in which the overlapping ends were annealed, allowing the 38 overlap of each strand to serve as a primer for the 38 extension of the complementary strand. The resulting fusion product was further amplified by PCR. This PCR product was inserted into pSP72 and its nucleotide sequence was confirmed using the 373A DNA sequencing system. Subsequently, the following three fragments were prepared: the first part of pCAG with the first part of hGMCSFR␣, the chimeric fragment from the latter part of hGM-CSFR␣ plus the first part of the transmembrane/intracellular mLIFR, and the latter part of transmembrane/intracellular mLIFR plus the latter part of pCAG. These three fragments were ligated, resulting in completion of the chimeric receptor under control of the CAG promoter. For constructing pMKIT-hGM-CSFRc-LIFR, the fragment of hGM-CSFRc-LIFR with the CAG promoter was isolated and ligated at the Bam HI and Not I sites of pMKIT neo, which was kindly provided by Dr K. Maruyama (Tokyo Medical and Dental University, Tokyo, Japan). Mutant receptors with progressive C-terminal truncations or cytoplasmic domain tyrosine = phenylalanine substitutions were constructed (see Fig 2). Chimeric cDNAs were subcloned into the Xho I site of pGEM-7Zf (Promega). C-terminal truncations were generated by PCR using a 58oligonucleotide (MDR1F: 58-CTGTAAGGCGCTACAGTTTCAGAA-38) located upstream of the unique Bgl II site in LIFR cDNA, and 38 oligonucleotides introducing termination codons and Bst EII sites (T112: 58-ATGGTGACCTACTGCACATCGATGTACACC-38; T129: 58-ATGGTGACCTACTCTGCTTTGGCTTGCGGC38; and T149: 58-ATGGTGACCTAGGGAAGGCGCATCTGTGG-38). Receptor fragments with tyrosine = phenylalanine substitutions were generated by recombinant PCR24 using MDR1F and MDR5R (58GACAAAGGGTGACCTGGTTA-38, which includes the normal termination codon and a Bst EII site) as external primers. The following oligonucleotides were used as internal primers: F5F, 58-GTGGCAGGCTTTAAGCCACAG-38; F5R, 58-CTGTGGCTTAAAGCCTGCCAC-38; F4F, 58-CAGTCCATGTTTCAGCCGCAA-38; F4R, 58-TTGCGGCTGAAACATGGACTG-38; F3F, 58-CAGGTGGTGTTCATCGATGTG-38; F3R, 58CACATCGATGAACACCACCTG-38; F6F, 58-ACCGCCGGTTTCAGACCTCAG-38; and F6R, 58-CTGAGGTCTGAAACCGGCGGT-38. Construct F3/4/5/6 was generated by recombination between F3 plasmid DNA and a fragment of F4/5/6 digested with Cla I and Bst EII. Similarly, F4/5/6 was generated by recombination between F4 and F5/6 1935 at the Bam HI and Bst EII sites. F5/6 was generated by PCR using F5 plasmid DNA as a template, and F6F and F6R as primers. The nucleotide sequences of the fragments derived on the PCR were confirmed by dideoxy sequencing using an ALF DNA sequencer (Pharmacia, Uppsala, Sweden). The construct of dominant negative STAT325 was kindly provided by Dr Alice L.-F.Mui (DNAX Research Institute, Palo Alto, CA). Transfection of DNA. DNA was introduced into cells by electroporation using a Gene Pulser (Bio-Rad, Richmond, CA). The cells (107) were suspended in 0.8 mL of HEPES-buffered saline (50 mmol/L HEPES, 137 mmol/L NaCl, 6.8 mmol/L KCl, 0.28 mmol/L Na2 HPO4, and 0.1% dextrose, pH 7.1) containing 25 µg each of the expression plasmids for chimeric GM-CSFR ␣ and c. The cells were exposed to a 450 V (WEHI-3B D⫹) or 400 V (M1) pulse with a capacitance of 960 µF. After 2 days culture, the cells were transferred to 96-well plates and then cultured in medium containing G-418 at the final concentration of 1.8 mg/mL. Surface expression of the transfected chimeric receptor genes were analyzed by flow cytometry with an Epics XL (Coulter Electronics, Luton, UK), using anti–GM-CSFR␣ (S-20; Santa Cruz Biotechnology, Santa Cruz, CA) and anti–GM-CSFR c (S-16; Santa Cruz Biotechnology). Transfectants expressing W238 receptors were further transfected with 30 µg of pME18S ⌬STAT3B together with 1 µg of pPUR (Clontech, Palo Alto, CA), which carries the puromycin-resistant gene. The cells were selected in a medium containing 2 µg/mL puromycin and 1.8 mg/mL G418. The C-terminal truncated STAT3 protein was immunoprecipitated using anti-STAT3 antibody K15 (directed against amino acids 626-640), which was obtained from Santa Cruz Biotechnology. Properties of differentiated cells. The differentiation of M1 cells was assayed by measuring the induction of phagocytic activity in the cells. M1 cells (3 ⫻ 105 cells/mL) were incubated with cytokines for 2 days. The cells were harvested, suspended in serum-free medium containing 0.2% of a suspension of polystyrene latex particles (average diameter, 0.944 µm, Seradyn, Indianapolis, IN), and then incubated for 4 hours at 37°C. The cells were then thoroughly washed three times with phosphate-buffered saline (PBS). Cells containing more than 10 latex particles were scored as phagocytic cells. The ability of cells to reduce nitroblue tetrazolium (NBT) was used as a functional marker of the differentiation of WEHI-3B D⫹ cells. The cells (104 cells/mL) were incubated with cytokines for 4 days. The cells were harvested, and the cell numbers were determined using a Model ZM Coulter Counter. The cells were suspended in 1 mL of serum-free medium containing 1 mg/mL of NBT and 100 ng/mL of 12-O-tetra decanoylphorbol 13-acetate, and then incubated for 1 hour at 37°C. The reaction was stopped by adding HCl to the final concentration of 1 mol/L. The suspension was centrifuged, and the formazan deposits were solubilized by adding dimethyl sulfoxide. The absorption of the formazan solution at 560 nm was measured with a spectrophotometer. Agar cultures of WEHI-3B D⫹ cells were performed in 35-mm Petri dishes, as described previously.26 Briefly, 200 cells were cultured in 1 mL of 0.3% agar (DIFCO Laboratories, Detroit, MI) in Dulbecco’s modified Eagle’s medium containing 20% FCS and various concentrations of hGM-CSF or IL-6 (100 ng/mL) as a positive inducer of differentiation. Colony numbers and morphology were scored after 7 days incubation at 37°C under a fully humidified atmosphere of 5% CO2 in air. Aggregates of more than 50 cells were scored as colonies. Colonies with a halo of dispersed cells were scored as differentiated. Immunoprecipitation and Western blot analysis. WEHI-3B D⫹ cells (107) were incubated with or without hGM-CSF (100 ng/mL) for 10 minutes at 37°C. The reaction was stopped by adding ice-cold PBS. The cells were harvested, washed once with ice-cold PBS, and then lysed in 400 µL of lysis buffer (1% Triton X-100, 150 mmol/L NaCl, 20 mmol/L Tris-HCl, pH 7.4, 2 mmol/L EDTA, 1 mmol/L Na3VO4, 1 mmol/L phenylmethylsulfonyl fluoride, 10 µg/mL leupeptin, 10 µg/mL pepstatin, 1 µg/mL aprotinin, and 100 µg/mL Pefabloc) for 5 From www.bloodjournal.org by guest on April 18, 2017. For personal use only. 1936 TOMIDA, HEIKE, AND YOKOTA Table 1. Differentiation of M1 Cells Expressing GM-CSFR␣-LIFR and GM-CSFR-LIFR Cytokine (ng/mL) Phagocytic Cells (%) None GM-CSF GM-CSF GM-CSF LIF IL-6 2.5 5 10 2 10 1.3 ⫾ 0.7* 27.5 ⫾ 9.0 36.8 ⫾ 4.3 48.0 ⫾ 6.0 22.8 ⫾ 4.8 39.4 ⫾ 7.6 Cells were incubated with cytokines for 2 days. Phagocytic activity of the cells was assayed as described in Materials and Methods. *Each value is the mean of duplicate determinations ⫾ standard error. RESULTS Signal transduction through the chimeric receptor carrying the full-length cytoplasmic domain of LIFR in M1 and WEHI-3B D⫹ cells. Stimulation of M1 cells with LIF or IL-6 induces macrophage differentiation and growth arrest.1,2,27-29 Although wild-type WEHI-3B D⫹ cells respond to IL-6, but not LIF, cells transfected with LIFR cDNA respond to both LIF and IL-6.5 M1 and WEHI-3B D⫹ cells were transfected with both the hGMCSFR␣-LIFR and hGM-CSFRc-LIFR constructs, and expression of these receptors was determined by flow cytometric analysis (Fig 1). The differentiation of M1 cells was assessed by measuring the induction of phagocytic activity in the cells, as the induction of phagocytic activity in the cells was associated with the induction of other phenotypic markers of cell differentiation.1 M1 cells expressing both the ␣ and c chimeric receptors responded to hGM-CSF, as well as LIF and IL-6 (Table 1). The differentiation of WEHI-3B D⫹ cells was Fig 1. Flow cytometric analysis of chimeric receptor expression in M1 transformants. M1 cells were transfected with GM-CSFR␣-LIFR and GM-CSFR c-LIFR cDNA. The cells were incubated with anti–GMCSFR ␣ (B) or anti–GM-CSFR c (C). Antibody binding was detected by incubation with fluorescein-conjugated rabbit antimouse immunoglobulin. Samples were analyzed using a flow cytometer. (A) The flow cytometry profile stained with the fluorescein-conjugated second antibody alone. minutes on ice. Cell lysates were centrifuged for 5 minutes at 4°C at 14,000 rpm, and the resulting supernatants were incubated with antibodies to STAT3 (C-20), STAT5b (C-17), SHP-2 (Santa Cruz Biotechnology), or Shc (Transduction Laboratories, Lexington, KY) for 2 hours at 4°C. Protein A-Sepharose was added to the reaction mixture, and the incubation was continued overnight at 4°C. The beads were pelleted by centrifugation and then washed three times with cold PBS. The immune complexes were eluted with 20 µL of Laemmli sample buffer, resolved by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and then transferred to Immobilon P (Millipore, Bedford, MA). The membrane was incubated with antiphosphotyrosine antibodies (4G10; Upstate Biotechnology, Lake Placid, NY) for 1 hour. After washing, the membrane was incubated with horseradish peroxidase-conjugated secondantibodies for 1 hour. The immune complex was detected using an enhanced chemiluminescence system (ECL; Amersham Life Science, Little Chalfont, UK). Blots were stripped with 2% SDS, 100 mmol/L 2-mercaptothanol, and 62.5 mmol/L Tris-HCl, reblocked, and then reprobed with antibodies to STAT3 (Transduction Laboratories). Fig 2. Deletion and point mutations in LIFR. The cytoplasmic regions of the LIFR mutants are schematically shown. The transmembrane domain (TM), putative box 1 (B1), box 2 (B2), and box 3 (B3), and the positions of tyrosine (Y) and phenylalanine (F) residues are indicated. From www.bloodjournal.org by guest on April 18, 2017. For personal use only. ROLE OF STAT3 IN DIFFERENTIATION OF WEHI-3B 1937 Fig 3. Differentiation of WEHI-3B Dⴙ cells expressing LIFR mutants. Cells expressing various chimeric receptors were treated for 4 days with 10 ng/mL of hGM-CSF or 100 ng/mL of IL-6. The NBT-reducing activity of the cells was determined by the colorimetric assay. The data for representative clones are presented as percentages of the values for untreated control cultures. The values represent the averages of duplicate assays ⴞ standard error. assessed first by measuring the ability of the cells to reduce NBT, a differentiation marker. The cells expressing both the ␣ and c chimeric receptors exhibited enhanced ability to reduce NBT on treatment with hGM-CSF, as well as IL-6 (see W238 in Fig 3). Cell migration and growth arrest of the transfected cells were also induced by treatment with hGM-CSF in a soft agar culture (see W238 in Fig 4). These results suggest that homodimerization of the LIFR cytoplasmic domain is sufficient to induce macrophage differentiation of M1 and WEHI-3B D⫹ cells because hGM-CSF cannot bind to mouse GM-CSFR. Cytoplasmic domains that are required for inducing differentiation and growth arrest of WEHI-3B D⫹ cells. To determine the receptor domains required for growth arrest and differentiation, LIFR mutants, as shown in Fig 2, were constructed. ␣ and c chimeric receptor cDNAs carrying these LIFR mutations were transfected into WEHI-3B D⫹ cells. hGM-CSF could induce differentiation and growth arrest of the transformants expressing truncated forms of the LIFR cytoplasmic domain containing at least 149 amino acid residues (Figs 3 and 4). More extensively truncated forms of the LIFR (T129 and T119) were unable to generate signals for differentiation and cell growth arrest. A normal differentiation response to IL-6 was observed in WEHI-3B D⫹ cells expressing each of the various mutant receptors. The phosphorylation of tyrosine residues is considered to be a crucial process in the initiation of signal transduction from the receptors after ligand binding. LIFR has six tyrosine residues in its cytoplasmic domain (Fig 2). The third tyrosine residue from the membrane-proximal region is located in a YXXV motif, a known docking site for SHP-2. Three tyrosine residues, ie, the fourth, fifth, and sixth ones, are located in a YXXQ motif, a STAT3 consensus binding sequence.18 We introduced a mutation into either the third (F3), fourth (F4), or fifth (F5) tyrosine residue, or into four tyrosine residues, ie, the third, fourth, fifth, and sixth ones (F3/4/5/6). Mutants, F3, F4, and F5, but not F3/4/5/6, could induce growth arrest and macrophage differentiation (Figs 3 and 4). STAT3 activation in relation to growth arrest and differentiation. LIF and related cytokines activate a common set of Fig 4. Soft agar colony assaying of WEHI-3B Dⴙ cells expressing LIFR mutants. Cells expressing the various chimeric receptors (W238, T149, T129, T119, F3, F4, F5, or F3/4/5/6) were cultured in agar with increasing concentrations of hGM-CSF or IL-6 (100 ng/mL). The colony numbers (left panels) and the proportions of colonies containing differentiated cells (right panels) were determined after 7 days. The means of duplicate assays are shown. From www.bloodjournal.org by guest on April 18, 2017. For personal use only. 1938 TOMIDA, HEIKE, AND YOKOTA Fig 5. Tyrosine phosphorylation of STAT3 through the chimeric receptors. Cells expressing various chimeric receptors were either stimulated (ⴙ) with hGM-CSF (10 ng/mL) for 10 minutes or left unstimulated (-). STAT3 was immunoprecipitated from cell lysates using anti-STAT3 antibodies and then probed with either antiphosphotyrosine antibodies (upper panel) or anti-STAT3 antibodies (lower panel). The immune complex was visualized by chemiluminescence. signalling molecules including components of the JAK-STAT pathway and of the Ras-MAP kinase signalling cascade.9,11,12 To assess the ability of the receptor mutants to activate Shc and SHP-2 in the Ras-MAP kinase pathway, and STAT3 and STAT5, Western blot analysis was performed on lysates of stimulated and unstimulated WEHI-3B D⫹ cells. Shc, SHP-2, and STAT5 were already phosphorylated in all cell lines cultured in medium containing FCS. Stimulation with hGM-CSF did not cause an increase in the level of tyrosine phosphorylation of these proteins (data not shown). On the contrary, phosphorylation of STAT3 was not detected in unstimulated cells. Stimulation with hGM-CSF induced the tyrosine phosphorylation of STAT3 in the transformants of W238 , T149, F3, F4, and F5, but not in the transformants of T129, T119, and F3/4/5/6 (Fig 5). The results show that the tight correlation between STAT3 activation and induction of differentiation and growth arrest of the cells. Effect of dominant-negative STAT3 on growth arrest and differentiation. To examine the role of STAT3, a truncated STAT3 (⌬STAT3), which lacks the transactivation domain and acts as a dominant negative manner was generated.25 The construct was transfected with a puromycin-resistance selection marker into WEHI-3B D⫹ cells expressing W238 receptors. Transfectants that were resistant to both puromycin and neomycin were isolated. The responsiveness of the cells to IL-6 was first examined because dominant negative forms of STAT3 were shown to block the IL-6–mediated growth arrest and differentiation of M1 cells.28,29 Several clones exhibiting different sensitivities to IL-6 were selected, and the expression of truncated A STAT3 in these clones was examined by immunoblot analysis. The expression of truncated STAT3 correlated with a decrease in sensitivity to IL-6. To examine the activation of STAT3, cells were stimulated with IL-6 or hGM-CSF. In the cells overexpressing truncated STAT3 (Fig 6B), the tyrosine-phosphorylation of endogenous STAT3 in response to IL-6 was reduced and the response to hGM-CSF was almost completely blocked (Fig 6A). We examined the effect of expression of the mutant STAT3 protein on the induction of differentiation and growth arrest of WEHI-3B D⫹ cells. As shown in Figs 7 and 8, in the cells expressing dominant negative STAT3, the induction of differentiation and growth arrest by hGM-CSF or IL-6 was dramatically reduced relative to the control. These results suggest that STAT3 plays an essential role in the signals mediated by the LIFR homodimer. DISCUSSION LIFR is a member of the hematopoietin receptor family.20 It is structurally most similar to gp130, G-CSFR, IL-12 receptor, OSM receptor, and leptin receptor.12,30,31 Among these receptors, gp130 and G-CSFR have been extensively studied as to the cytoplasmic domains required for cell proliferation and differentiation.27-29,32,33 While these receptor subunits function as homodimers, LIFR functions as a heterodimer with gp130. To examine signal transduction by LIF, we fused the cytoplasmic domain of mouse LIFR or gp130 to the extracellular domain of the human GM-CSFR␣ and c chains, respectively. By transfection of different combinations of chimeric receptors into mouse B Fig 6. Inhibition of tyrosine phosphorylation of STAT3 by dominant negative STAT3. (A) WEHI-3B Dⴙ cells expressing the chimeric LIFR (W238) and ⌬STAT3 or the chimeric LIFR alone (PurrNeor) were stimulated with hGM-CSF (10 ng/mL) or IL-6 (100 ng/mL) for 10 minutes. STAT3 was immunoprecipitated from cell lysates using anti-STAT3 antibodies (C20; Santa Cruz Biotechnology), and then probed with either antiphosphotyrosine antibodies or anti-STAT3 antibodies (Transduction Laboratories). (B) Expression of ⌬STAT3 in WEHI-3B Dⴙ cells. STAT3 was immunoprecipitated from cell lysates using anti-STAT3 antibodies (K-15; Santa Cruz Biotechnology) and then probed with anti-STAT3 antibodies (Transduction Laboratories). From www.bloodjournal.org by guest on April 18, 2017. For personal use only. ROLE OF STAT3 IN DIFFERENTIATION OF WEHI-3B Fig 7. Inhibition of differentiation of WEHI-3B Dⴙ cells expressing LIFR by dominant negative STAT3. Cells expressing LIFR (W238) and ⌬STAT3, or LIFR alone (PurrNeor) were treated for 4 days with 10 ng/mL of hGM-CSF or 100 ng/mL of IL-6. The NBT-reducing activity of the cells was determined by the colorimetric assay. The data for representative clones are presented as percentages of the values for untreated control cultures. The values represent the averages of duplicate assays ⴞ standard error. cells, we analyzed the signals of not only homodimers, but also heterodimers of LIFR and gp130.39 In the present study, we transfected M1 and WEHI-3B D⫹ cells with GM-CSFR␣-LIFR and GM-CSFRc-LIFR cDNA and found that hGM-CSF induced macrophage differentiation and growth arrest of these leukemic cells, suggesting that the cytoplasmic domain of LIFR is capable of signal transduction Fig 8. Soft agar colony assaying of WEHI-3B Dⴙ cells expressing LIFR and dominant negative STAT3. Cells expressing LIFR (W238) and ⌬STAT3, or LIFR alone (PurrNeor) were cultured in agar with increasing concentrations of hGM-CSF or IL-6 (100 ng/mL). The colony numbers (left panels) and the proportions of colonies containing differentiated cells (right panels) were determined after 7 days. The means of duplicate assays are shown. 1939 when it is induced to form a homodimer. Therefore, mutants of these chimeric receptors permit analysis of the functional domain of the LIFR cytoplasmic domain in the absence of gp130. We identified the first 149 amino acid residues of the cytoplasmic domain of LIFR as the minimal region necessary for growth arrest and differentiation of WEHI-3B D⫹ cells. The amino acid 136-145 region, called box 3, is conserved among gp130, LIFR and G-CSFR and contains the fifth tyrosine residue and a YXXQ motif. The tyrosine residue in the motif was shown to play a role in activating STAT3.18 The fourth and sixth tyrosine residues are also contained in the YXXQ motif.21 However, the truncated mutant, T129, containing the fourth tyrosine residue and a YXXQ motif, neither activated STAT3 nor induced the differentiation of WEHI-3B D⫹ cells. The mutant, T149, with the F5 mutation was also inactive (data not shown). Therefore, the sixth tyrosine residue may substitute for the fifth tyrosine residue in activating STAT3 and the generation of the signals for growth arrest and differentiation because the F5 mutation in the full-length cytoplasmic domain did not affect the function of the receptor (Figs 3 and 4). Kuropatwinski et al19 and Stahl et al18 fused the cytoplasmic domain of LIFR and the extracellular domains of G-CSFR and the epidermal growth factor receptor, respectively. These chimeric receptors formed homodimers after binding to ligands. They showed that similar constructs to our T129 mutant could activate STAT3 in COS-1 or COS-7 cells. The difference between their and our present results seems to originate from the difference between the lineages of the cells used. Our present study showed that activation of STAT3 is tightly correlated with the signals for growth arrest and macrophage differentiation, and that a dominant negative form of STAT3 inhibited both LIFR-mediated growth arrest and macrophage differentiation of the WEHI-3B D⫹ transformants. These results are consistent with the results obtained by Nakajima et al28 and Minami et al.29 They showed that activation of STAT3 is essential for gp130-mediated growth arrest and differentiation of M1 cells. On the other hand, Alexander et al34 showed that activation of STAT3 DNA binding was insufficient for c-Mpl From www.bloodjournal.org by guest on April 18, 2017. For personal use only. 1940 TOMIDA, HEIKE, AND YOKOTA (thrombopoietin receptor)-mediated WEHI-3B D⫹ differentiation. They suggested that activation of components of the Ras signalling cascade, initiated by interaction of Shc with c-Mpl, plays a decisive role in the differentiation signals. Therefore, we examined Shc phosphorylation in our WEHI-3B D⫹ cells. In contrast to their results, Shc was phosphorylated in the cells cultured in normal medium containing FCS. Treatment of the cells with hGM-CSF did not cause a further increase in phosphorylation. Nicholson et al33 analyzed tyrosine residues in G-CSFR that mediate the induction of differentiation of M1 cells. They showed that STAT3 was activated by G-CSF in the cells expressing G-CSFR tyrosine mutants unable to mediate G-CSF–induced differentiation. Therefore, activation of STAT3 is not sufficient for G-CSF–mediated M1 differentiation. Tyrosine phosphorylation of Shc was not required for the differentiation of M1 cells. All of these results, including ours, suggest that activation of STAT3 is required, although not sufficient, for induction of the differentiation of myeloid leukemic cells such as WEHI-3B D⫹ and M1. Multiple signaling pathways through LIF in various cells have been reported.35-37 Recently, Starr et al38 reported that homodimers of the LIFR cytoplasmic domain were able to deliver the signal that blocks differentiation of ES cells, although they were not sufficient for transducing a differentiation signal in M1 cells nor for induction of a proliferation signal in Ba/F3 cells. We also examined the effects of our chimeric receptors on ES and Ba/F3 cells.39 Homodimers of the LIFR cytoplasmic domain could not support the proliferation of these cells. The reason for the difference between our results and theirs is not known, but clonal variation of ES and M1 cells and differences in the selection of transfectants may be possible explanations. In general, LIFR homodimers may be less efficient than heterodimers of LIFR and gp130 in signaling biological responses. However, in the case of WEHI-3B D⫹ cells, LIFR homodimers were equivalent in activity to heterodimers of LIFR and gp130. The response to hGM-CSF of WEHI-3BD⫹ cells transfected with the present chimeric receptors was very similar to the response to LIF of WEHI-3B D⫹ cells transfected with the full-length LIFR, which formed a heteromeric complex with endogenous gp130.5 We showed that the cytoplasmic domain of LIFR in the absence of gp130 could generate the signals for growth arrest and differentiation of myeloid leukemic cells. It is possible that an as yet undiscovered cytokine could induce downstream signaling through homodimerization of LIFR in some cell types. No signaling pathway specific to LIFR has been identified. Our chimeric receptors will be useful for elucidating the different signaling potentials of LIFR versus gp130. ACKNOWLEDGMENT We thank T. Higashihara for her technical assistance and Dr T. Matsuda for the helpful discussions. REFERENCES 1. Tomida M, Yamamoto-Yamaguchi Y, Hozumi M: Purification of a factor inducing differentiation of mouse myeloid leukemic M1 cells from conditioned medium of mouse fibroblast L929 cells. J Biol Chem 259:10978, 1984 2. Tomida M, Yamamoto-Yamaguchi Y, Hozumi M, Okabe T, Takaku F: Induction by recombinant human granulocyte colonystimulating factor of differentiation of mouse myeloid leukemic M1 cells. FEBS Lett 207:271, 1986 3. Gearing DP, Gough NM, King JA, Hilton DJ, Nicola NA, Simpson RJ, Nice EC, Kelso A, Metcalf D: Molecular cloning and expression of cDNA encoding a murine myeloid leukaemia inhibitory factor (LIF). EMBO J 6:3995, 1987 4. Lowe DG, Nunes W, Bombara M, McCabe S, Ranges GE, Henzel W, Tomida M, Yamamoto-Yamaguchi Y, Hozumi M, Goeddel DV: Genomic cloning and heterologous expression of human differentiationstimulating factor. DNA 8:351, 1989 5. Tomida M: Induction of differentiation of WEHI-3B D⫹ leukemic cells transfected with differentiation-stimulating factor/leukemia inhibitory factor receptor cDNA. Blood 85:217, 1995 6. Gearing DP: The leukemia inhibitory factor and its receptor. Adv Immunol 53:31, 1993 7. Alexander HR, Billingsley KG, Block MI, Fraker DL: D-factor/ leukaemia inhibitory factor: Evidence for its role as a mediator in acute and chronic inflammatory disease. Cytokine 6:589, 1994 8. Baumann H, Symes AJ, Comeau MR, Morella KK, Wang Y, Friend D, Ziegler SF, Fink JS, Gearing DP: Multiple regions within the cytoplasmic domains of the leukemia inhibitory factor receptor and gp130 cooperate in signal transduction in hepatic and neuronal cells. Mol Cell Biol 14:138, 1994 9. Ernst M, Oates A, Dunn AR: gp130-mediated signal transduction in embryonic stem cells involves activation of Jak and Ras/mitogenactivated protein kinase pathways. J Biol Chem 271:30136, 1996 10. Pennica D, Shaw KJ, Swanson TA, Moore MW, Shelton DL, Zioncheck KA, Rosenthal A, Taga T, Paoni NF, Wood WI: Cardiotrophin-1: Biological activities and binding to the leukemia inhibitory factor receptor/gp130. J Biol Chem 270:10915, 1995 11. Sheng Z, Knowlton K, Chen J, Hoshijima M, Brown JH, Chien KR: Cardiotrophin 1 (CT-1) inhibition of cardiac myocyte apoptosis via a mitogen-activated protein kinase-dependent pathway: Divergence from downstream CT-1 signals for myocardial cell hypertrophy. J Biol Chem 272:5783, 1997 12. Kishimoto T, Akira S, Narazaki M, Taga T: Interleukin-6 family of cytokines and gp130. Blood 86:1243, 1995 13. Ware CB, Horowitz MC, Renshaw BR, Hunt JS, Liggitt D, Koblar SA, Gliniak BC, Mckenna HJ, Papayannopoulou T, Thoma B, Cheng L, Donovan P J, Peschon JJ, Bartlett PF, Willis CR, Wright BD, Carpenter MK, Davison BL, Gearing DP: Targeted disruption of the low-affinity leukemia inhibitory factor receptor gene causes placental, skeletal, neural and metabolic defects, and results in perinatal death. Development 121:1283, 1995 14. Li M, Sendtner M, Smith A: Essential function of LIF receptor in motor neurons. Nature 378:724, 1995 15. Miyajima A, Mui AL-F, Ogorochi T, Sakamaki K: Receptors for granulocyte-macrophage colony-stimulating factor, interleukin-3, and interleukin-5. Blood 82:1960, 1993 16. Ihle JN, Kerr IM: Jaks and Stats in signaling by the cytokine receptor superfamily. Trends Genet 11:69, 1995 17. Darnell JE Jr, Kerr IM, Stark GR: Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264:1415, 1994 18. Stahl N, Farruggella TJ, Boulton TG, Zhong Z, Darnell JE Jr, Yancopoulos GD: Choice of STATs and other substrates specified by modular tyrosine-based motifs in cytokine receptors. Science 267:1349, 1995 19. Kuropatwinski KK, De Imus C, Gearing D, Baumann H, Mosley B: Influence of subunit combinations on signaling by receptors for oncostatin M, leukemia inhibitory factor, and interleukin-6. J Biol Chem 272:15135, 1997 From www.bloodjournal.org by guest on April 18, 2017. For personal use only. ROLE OF STAT3 IN DIFFERENTIATION OF WEHI-3B 20. Gearing DP, Thut CJ, VandenBos T, Gimpel SD, Delaney PB, King J, Price V, Cosman D, Beckmann MP: Leukemia inhibitory factor receptor is structurally related to the IL-6 signal transducer, gp130. EMBO J 10:2839, 1991 21. Tomida M, Yamamoto-Yamaguchi Y, Hozumi M: Three different cDNAs encoding mouse D-factor/LIF receptor. J Biochem 115:557, 1994 22. Niwa H, Yamamura K, Miyazaki J: Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193, 1991 23. Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR: Sitedirected mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51, 1989 24. Higuchi R: Recombinant PCR, in Innis MA, Gelfand DH, Shinsky JJ, White TJ (eds): PCR Protocols. San Diego, CA, Academic, 1990, p 177 25. O’Farrell A-M, Liu Y, Moore KW, Mui AL-F: IL-10 inhibits macrophage activation and proliferation by distinct signaling mechanisms: Evidence for Stat3-dependent and -independent pathways. EMBO J 17:1006, 1998 26. Metcalf D: The Hemopoietic Colony Stimulating Factors. Amsterdam, The Netherlands, Elsevier, 1984 27. Yamanaka Y, Nakajima K, Fukada T, Hibi M, Hirano T: Differentiation and growth arrest signals are generated through the cytoplasmic region of gp130 that is essential for Stat3 activation. EMBO J 15:1557, 1996 28. Nakajima K, Yamanaka Y, Nakae K, Kojima H, Ichiba M, Kiuchi N, Kitaoka T, Fukada T, Hibi M, Hirano T: A central role for Stat3 in IL-6-induced regulation of growth and differentiation in M1 leukemia cells. EMBO J 15:3651, 1996 29. Minami M, Inoue M, Wei S, Takeda K, Matsumoto M, Kishimoto T, Akira S: STAT3 activation is a critical step in gp130-mediated terminal differentiation and growth arrest of a myeloid cell line. Proc Natl Acad Sci USA 93:3963, 1996 30. Mosley B, De Imus C, Fried D, Boiani N, Thoma B, Park LS, Cosman D: Dual oncostatinM(OSM) receptors. Cloning and characterization of an alternative signaling subunit conferring OSM-specific receptor activation. J Biol Chem 271:32635, 1996 1941 31. Tartaglia LA, Dembski M, Weng X, Deng N, Culpepper J, Devos R, Richards GJ, Campfield LA, Clark FT, Deeds J, Muir C, Sanker S, Moriarty A, Moore KJ, Smutko JS, Mays GG, Woolf EA, Monroe CA, Tepper RI: Identification and expression cloning of a leptin receptor, OB-R. Cell 83:1263, 1995 32. Yoshikawa A, Murakami H, Nagata S: Distinct signal transduction through the tyrosine-containing domains of the granulocyte colonystimulating factor receptor. EMBO J 14:5288, 1995 33. Nicholson SE, Starr R, Novak U, Hilton DJ, Layton JE: Tyrosine residues in the granulocyte colony-stimulating factor (G-CSF) receptor mediate G-CSF-induced differentiation of murine myeloid leukemic (M1) cells. J Biol Chem 271:26947, 1996 34. Alexander WS, Maurer AB, Novak U, Harrison-Smith M: Tyrosine-599 of the c-Mpl receptor is required for Shc phosphorylation and the induction of cellular differentiation. EMBO J 15:6531, 1996 35. Boulton TG, Stahl N, Yancopoulos GD: Ciliary neurotrophic factor/leukemia inhibitory factor/interleukin 6/oncostatin M family of cytokines induces tyrosine phosphorylation of a common set of proteins overlapping those induced by other cytokines and growth factors. J Biol Chem 269:11648, 1994 36. Schiemann WP, Nathanson NM: Involvement of protein kinase C during activation of the mitogen-activated protein kinase cascade by leukemia inhibitory factor. Evidence for participation of multiple signaling pathways. J Biol Chem 269:6376, 1994 37. Yin T, Yang Y: Mitogen-activated protein kinases and ribosomal S6 protein kinases are involved in signaling pathways shared by interleukin-11, interleukin-6, leukemia inhibitory factor, and oncostatinMin mouse 3T3-L1 cells. J Biol Chem 269:3731, 1994 38. Starr R, Novak U, Willson TA, Inglese M, Murphy V, Alexander WS, Metcalf D, Nicola NA, Hilton DJ, Ernst M: Distinct roles for leukemia inhibitory factor receptor ␣-chain and gp130 in cell typespecific signal transduction. J Biol Chem 272:19982, 1997 39. Nakamura T, Arai T, Takagi M, Sawada T, Matsuda T, Yokota T, Heike T: A selective switch-on system for self-renewal of embryonic stem cells using chimeric cytokine receptors. Biochem Biophys Res Commun 248:22, 1998 From www.bloodjournal.org by guest on April 18, 2017. For personal use only. 1999 93: 1934-1941 Cytoplasmic Domains of the Leukemia Inhibitory Factor Receptor Required for STAT3 Activation, Differentiation, and Growth Arrest of Myeloid Leukemic Cells Mikio Tomida, Toshio Heike and Takashi Yokota Updated information and services can be found at: http://www.bloodjournal.org/content/93/6/1934.full.html Articles on similar topics can be found in the following Blood collections Hematopoiesis and Stem Cells (3419 articles) Information about reproducing this article in parts or in its entirety may be found online at: http://www.bloodjournal.org/site/misc/rights.xhtml#repub_requests Information about ordering reprints may be found online at: http://www.bloodjournal.org/site/misc/rights.xhtml#reprints Information about subscriptions and ASH membership may be found online at: http://www.bloodjournal.org/site/subscriptions/index.xhtml Blood (print ISSN 0006-4971, online ISSN 1528-0020), is published weekly by the American Society of Hematology, 2021 L St, NW, Suite 900, Washington DC 20036. Copyright 2011 by The American Society of Hematology; all rights reserved.