* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download A Connective Tissue Disorders NGS Panel: Development
Saethre–Chotzen syndrome wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Point mutation wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Public health genomics wikipedia , lookup
Gene therapy wikipedia , lookup
Frameshift mutation wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Gene expression profiling wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Pharmacogenomics wikipedia , lookup
Genome (book) wikipedia , lookup
Medical genetics wikipedia , lookup
Down syndrome wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Microevolution wikipedia , lookup
Neuronal ceroid lipofuscinosis wikipedia , lookup
DiGeorge syndrome wikipedia , lookup
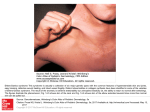
A Connective Tissue Disorders NGS Panel: Development, Validation, and Novel Findings Jennifer A. Lee, Monica J. Basehore, Stephen McGee, Katharine Kubiak, Kellie King, Kristen J. Champion, Julie R. Jones, and Michael J. Friez www.GGC.org Greenwood Genetic Center, Greenwood, South Carolina 29646, USA Background Diagnostic Workflow Connective tissues are the structurally supportive components of the body that form a framework or structural matrix, connect body tissues, and provide protection of organs and storage of energy. Connective tissue disorders represent a heterogeneous group of more than 200 recognized conditions for which the connective tissues are the primary pathologic target. These disorders fall into two categories: autoimmune and heritable. The heritable disorders affect a wide variety of mesenchymal tissues, and include either soft connective tissue disorders which may be characterized by excessive skin laxity, joint hypermobility, easy bruising, or skeletal dysplasias which may affect the development of the bones. Making a specific diagnosis can be important in determining the appropriate medical management. Many of these conditions can have life-threatening complications (i.e., aortic root rupture, bowel rupture, etc.), many require regular monitoring/surveillance of symptoms, and for many of these conditions treatment is available. While certain connective tissue disorders are associated with definitive phenotypes, some patients may have non-specific or atypical presentations. Also, many patients present with similar features due to the clinical variability and variable expressivity observed among these disorders. For cases of phenotypic variability and variable expressivity, we have developed and validated a targeted sequencing panel for routine clinical diagnostic testing. Using RainDanceTM microdroplet enrichment and SOLiDTM Next Generation Sequencing (NGS), we can analyze the coding and flanking intronic regions of 31 genes (Table 1) associated with various connective tissue disorders simultaneously. This method enables the detection of single nucleotide changes and small duplications and deletions in genes that, collectively, are associated with at least 80 distinct connective tissue disorder phenotypes (Table 2). Connective Tissue 31-Gene Panel • ABCC6 • ACTA2 • ACVR1 • ADAMTS2 • ATP6V0A2 • CBS • CHST14 • COL11A1 • COL1A1 • COL1A2 • COL2A1 • COL3A1 • COL5A1 • COL5A2 • ELN • FBLN5 • FBN1 • FBN2 • FKBP14 • MYLK • NOTCH1 • PKD2 • PLOD1 • PRDM5 • SLC2A10 • SLC39A13 • SMAD3 • TGFBR1 • TGFBR2 • TNXB • ZNF469 Table 1. List of 31 connective tissue disorder genes on the NGS panel Upon reviewing the current literature, 31 genes were selected for inclusion in the Connective Tissue gene panel. Inheritance patterns include autosomal dominant and autosomal recessive. Detection and Limitations • Can detect single nucleotide changes and small duplications and deletions in genes that are associated with at least 80 distinct connective tissue disorder phenotypes • Large deletions/duplications (CNVs) not detected • 99% of targeted sequence can be analyzed: +/- 20 bp at intron/exon boundaries Promoters and 3’-UTRs are not included Indications for Clinical Testing • Patients with non-specific presentation who do not fit the typical phenotype for a specific connective tissue disorder • For patients with a specific suspected connective tissue disorder, individual gene sequencing should be considered first • Molecular testing is useful to confirm the diagnosis and to identify the disease causing mutations within a family to allow for carrier testing and prenatal diagnosis Patient 3 DNA extraction Enrichment for panel genes (RainDanceTM) Figure 3. Clinical signs observed for patient 3. Library Preparation & Emulsion PCR • For patient 3, differential diagnoses of brittle cornea syndrome and osteogenesis imperfecta were considered due to keratoglobus bilateral, blue/purple sclera, abnormal red reflex, no skin or cardiac findings (Figure 3). • Results: apparently homozygous c.6644delA in ZNF469 gene (frameshift, predicted pathogenic) • Mutation is maternal; also has paternal deletion including ZNF469 by array. • The ZNF469 gene encodes a zinc-finger protein. It may function as a transcription factor or extra-nuclear regulator factor for the synthesis or organization of collagen fibers. Mutations in this gene cause brittle cornea syndrome. • Diagnosis: brittle cornea syndrome (autosomal recessive) NGS (SOLiDTM) Sanger sequencing: RDOs Director analysis Sanger sequencing: confirmations & UDOs Director review & interpretation of all Sanger data Figure 1. Diagnostic workflow RDO : recurrent dropout UDO : unique dropout Reporting Molecular Methods DNA extraction was performed using standard methods. Five micrograms of each genomic DNA sample were fragmented to 4-6 kb using a Covaris instrument. Enrichment for the 31 connective tissue genes was performed using an RDT1000 instrument (RainDance TechnologiesTM). Standard fragment libraries were prepared for each sample, library amplification was performed using emulsion PCR, and these PCR products were purified (AMPure) and then deposited onto glass slides for analysis by the SOLiDTM 5500xl system (Applied Biosystems). After bioinformatic processing, these data were analyzed to identify novel alterations as well as those previously reported in the Human Gene Mutation Database (HGMD). Identified alterations for a given patient were cross-referenced to those found for other samples within the same run, as well as to a cumulative database of sample results from all previous runs. After filtering out known SNPs, we could determine which changes were most likely novel, thus warranting further consideration. All potentially clinically relevant changes were confirmed by Sanger sequencing using custom primers. In addition to NGS, a dropout Sanger sequencing panel was performed for each sample to analyze those regions that are missed routinely (recurrent dropout, RDO) or have poor coverage (unique dropout, UDO) using NGS. Marfan syndrome Marshall syndrome MASS syndrome Moyamoya disease Osteogenesis Imperfecta Polycystic kidney disease (AD) Pseudoxanthoma elasticum Shprintzen-Goldberg syndrome Stickler syndrome Stiff skin syndrome Spondylocheirodysplasia Spondyloperipheral dysplasia Supravalvular aortic stenosis Thoracic aortic aneurysms & dissections Wrinkly skin syndrome and more… Table 2. List of connective tissue disorders associated with the 31 interrogated genes • Patient 4 has the following signs: impressive joint hypermobility, irregular menses, bilateral carpal tunnel syndrome, mild scoliosis, and postural orthostatic tachycardia syndrome. • Results: heterozygous c.2539C>T (p.R847X) in TNXB gene (nonsense, predicted pathogenic) • The TNXB gene encodes a member of the tenascin family of extracellular matrix glycoproteins. The tenascins have anti-adhesive effects, as opposed to fibronectin which is adhesive. This protein is thought to function in matrix maturation during wound healing, and its deficiency has been associated with Ehlers-Danlos syndrome. • Diagnosis: Ehlers-Danlos syndrome hypermobility type (autosomal dominant) Patient 5 Preliminary Results Molecular testing was performed using our diagnostic NGS panel on an initial cohort of 22 patients with suspected connective tissue disorders. Since the submission of this abstract, an additional 24 patients had been tested. Of all 46 patients tested to date, we identified novel likelypathogenic changes in five (11%) of these patients (see below, Figures 2-4). Of the remaining patients, only 6 (13%) were found to have normal results with no potentially pathogenic molecular alterations detected, and 35 (76%) had at least one variant of uncertain clinical significance (VUS). Patient 1 • Patient 1 has features of osteogenesis imperfecta: easy fractures. • Results: heterozygous c.2550delT in COL1A1 gene (frameshift, predicted pathogenic) + 4 VUS • The COL1A1 gene encodes the pro-alpha1 chains of type I collagen. Type I is a fibril-forming collagen found in most connective tissues and is abundant in bone, cornea, dermis, and tendon. Mutations in this gene are associated with osteogenesis imperfecta types I-IV, Ehlers-Danlos syndrome type VIIA and classical type, Caffey Disease, and idiopathic osteoporosis. • Diagnosis: osteogenesis imperfecta (autosomal dominant) Patient 2 Associated Disorders Achondrogenesis Aortic valve disease Arterial tortuosity syndrome Arthrogryposis Beals contractural arachnodactyly Brittle cornea syndrome Caffey disease Cutis laxa Czech dysplasia Ehlers-Danlos syndrome Epiphyseal dysplasia Fibrochondrogenesis Fibrodysplasia ossificans progressiva Homocystinuria Hypochondrogenesis Legg-Calve-Perthes disease Loeys-Dietz syndrome Patient 4 Figure 2. Clinical signs observed for patient 2. • Patient 2 has features of Ehlers-Danlos syndrome classical or type II: easy bruising/abnormal wound healing, joint hypermobility, soft skin (Figure 2); her father has similar signs: hyperextensible elbows, skin elasticity, problems with wound healing and scarring. • Results: heterozygous c.4714delG in COL5A1 gene (frameshift, predicted pathogenic) + 1 VUS • The COL5A1 gene encodes alpha chain of a low abundance fibrillar collagen. Type V collagen is found in tissues containing type I collagen and appears to regulate the assembly of heterotypic fibers composed of both type I and type V collagen. Mutations in this gene are associated with Ehlers-Danlos syndrome types I and II. • Diagnosis: Ehlers-Danlos syndrome type I or II (autosomal dominant) Figure 4. Clinical signs observed for patient 5. • Patient 5 has features of Ehlers-Danlos syndrome type I or other CT disorder: joint hypermobility, patellar dislocations, flat feet, no skin or cardiac findings, upslanting palpebral fissures, hypertelorism, low-set posteriorly rotated ears, mild brachyclinodactyly of 5th fingers, and squaring of the thumbs (Figure 4). • Results: heterozygous c.3527-1G>T in COL1A2 gene (splice site, predicted pathogenic) + 3 VUS • The COL1A2 gene encodes the pro-alpha2 chain of type I collagen. Type I is a fibril-forming collagen found in most connective tissues and is abundant in bone, cornea, dermis and tendon. Mutations in this gene are associated with osteogenesis imperfecta types I-IV, Ehlers-Danlos syndrome type VIIB, recessive Ehlers-Danlos syndrome classical type, idiopathic osteoporosis, and atypical Marfan syndrome. • Diagnosis: Ehlers-Danlos syndrome type VIIB (autosomal dominant) Summary • For cases of phenotypic variability and variable expressivity, we have developed and validated a targeted sequencing panel for routine clinical diagnostic testing. • With our Connective Tissue NGS panel, we analyze coding and flanking intronic regions of 31 genes associated with various connective tissue disorders simultaneously. • We can detect single nucleotide changes and small duplications and deletions in genes that are associated with at least 80 distinct connective tissue disorder phenotypes. • Preliminary findings suggest that our targeted NGS panel may have a potentially high diagnostic value for patients with suspected connective tissue disorders. • Our mutation detection rate is approximately 11%, and is expected to increase as more family members are tested to clarify the clinical significance of detected unclassified variants. • The simultaneous analysis of a panel of 31 connective tissue disorder genes enables a more efficient diagnostic strategy compared to a singlegene approach using traditional Sanger sequencing. Acknowledgments We thank the clinicians and genetic counselors who have referred patients for clinical diagnostic testing. We also recognize the contributions of our laboratory staff for their efforts to provide comprehensive clinical testing.