* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Analysis of the glycoside hydrolase family 8 catalytic core in
Magnesium transporter wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Western blot wikipedia , lookup
Biosynthesis wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Catalytic triad wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Point mutation wikipedia , lookup
Genetic code wikipedia , lookup
Biochemistry wikipedia , lookup
Metalloprotein wikipedia , lookup
Proteolysis wikipedia , lookup
Two-hybrid screening wikipedia , lookup
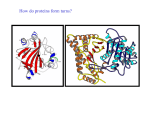
International Journal of Computational Bioinformatics and In Silico Modeling Vol. 3, No. 1 (2014): 315-320 Research Article Open Access ISSN: 2320-0634 Analysis of the glycoside hydrolase family 8 catalytic core in cellulase-chitosanases from Bacillus species Niveditha Prakash and Shubha Gopal* Department of studies in Microbiology, University of Mysore, Manasagangothri, Mysore, Karnataka, India *Corresponding author: Shubha Gopal; e-mail: [email protected] Received: 01 December 2013 Accepted: 12 December 2013 Online: 03 January 2014 ABSTRACT The glycoside hydrolase family 8 (GH-8) consists of bifunctional cellulase-chitosanases many of which are produced by species of Bacillus. Chitosanolytic enzymes can be useful in producing low molecular weight chitooligosaccharides which have several applications. In addition, a bifunctional enzyme would be more beneficial than the use of two individual enzymes in the production of chitooligosaccharides and in the degradation of the complex cellulose, chitin and chitosan rich biomass that occurs in nature. The crystal structure of a previously determined GH-8 chitosanase has revealed that the catalytic site is constructed on a scaffold of a double α6/α6 barrel which consists of six helix-loop-helix motifs. Bifunctional cellulase-chitosanases from Bacillus species isolated hitherto were analysed and were found to have the double α6/α6 barrel as previously predicted. The signature pattern of glycoside hydrolase family 8 (GH-8) was determined for the sequences. Some of the protein sequences among them were modelled and their closest structural analogs were determined. The structures were found to be related to endoglucanases, xylanases, epimerases and also terpenoid cyclases all of which have the double α6/α6 barrel architecture. This study provides a detailed insight into the structure of cellulase-chitosanase catalytic core and identifies related enzymes with similar catalytic core thereby signifying a possible evolutionary relationship. Keywords: Glycoside hydrolase family 8; Bacillus; six hairpin glycosidases; chitosanase-cellulases; homology modelling. INTRODUCTION Chitosan is hydrolysed by chitosanase [EC 3.2.1.132], an enzyme that catalyses the hydrolysis of glycosidic bonds of chitosan, the deacetylated derivative of chitin [1]. Chitosanases are classified under glycoside hydrolases, a diverse group of enzymes containing 93 families [2]. Bifunctional enzymes with cellulolytic and chitosanolytic activity mostly belong to glycoside hydrolase family 8 (GH-8) [3]. GH-8 is a chitosanase family which also has cellulase, xylanase, lichenase etc. Most of the bifunctional cellulase-chitosanases, particularly the ones produced by Bacillus species belong to glycoside hydrolase family 8. Their primary sequences show high homology with other glucanases of GH-8; xylanases, endoglucanases, lichenases and a few others [3]. Previously resolved crystal structure of GH-8 chitosanases catalytic domain from Bacillus sp. K17 by Adachi et al. [4] has revealed that the molecule consists of six α-helices forming a central barrel, http://bioinfo.aizeonpublishers.net/content/2014/1/bioinfo315-320.pdf surrounded by six other α-helices. Six repetitions of this helix-loop-helix motif form a typical α6/α6 double barrel structure. The structures have long loops with short β-strands and 310 helices protrude from the end of the outer helices and fold towards the inner helices forming a long cleft on the top of the double barrel suggestive of substrate binding pockets. On the bottom of the double barrel structure, short loops link the helices [4]. GH-8 chitosanases having the typical α6/α6 double barrel structure are called six hairpin glycosidases. The six-hairpin glycoside domain contains about seven alpha-hairpins arranged in closed circular assortment. In the present study, bifunctional cellulasechitosanases that have been previously reported are structurally examined for presence of the six hairpin glycoside domain. The protein structures are generated using online homology modelling servers. GH-8 signature motif is determined for the sequences, their active site residue is predicted and their closest 315 Niveditha Prakash and Shubha Gopal / Int J Comput Bioinfo In Silico Model. 2014, 3(1): 315-320 structural analogs are detected. The presence of the α6/α6 double barrel architecture in proteins from different families although their active-site architecture may be different suggests the stability of the α6/α6 motif and the probable evolutionary relationship between different glycosyl hydrolases [5]. MATERIALS AND METHODS Analysis of cellulase-chitosanase sequences from Bacillus species Previously reported protein sequences of Bacillus species showing cellulase and chitosanase activities were retrieved from NCBI [www.ncbi.nlm.nih.gov/protein] with the following accession numbers: Chitosanase from Bacillus cereus H1 (Q849S1) [6], Bacillus sp. strain KCTC 0377BP (QALZ1) [7] and cellulase-chitosanases from Bacillus thuringiensis subspecies (AB061887, AB061888, AB061889, AB061891, AB061892, AB061893, AB061894 and AB061895) [8]. Multiple sequence alignment and analysis of the glycosyl hydrolases family 8 signature motif The protein sequences were aligned using Clustal Omega set at default parameters [www.ebi.ac.uk/Tools/msa/clustalo] [9-10]. The Multiple sequence alignment output from Clustal Omega was obtained using Jalview [http://www.jalview.org/] [11]. The protein sequences were analysed with Prosite [http://prosite.expasy.org/scanprosite/] [12] and the signature motifs of the enzymes were obtained. The position of the signature motif in the multiple sequence alignment was detected. Sequence logo was generated using Weblogo [http://weblogo.berkeley.edu/logo.cgi] [13]. Homology modelling and identification of structural homologs Chitosanase-cellulase protein sequences of the above mentioned protein sequences were retrieved from NCBI [www.ncbi.nlm.nih.gov/protein]. Chitosanase from Bacillus cereus H1 (Q849S1) [6], Bacillus sp. strain KCTC 0377BP (QALZ1) [7] and one from Bacillus thuringiensis subspeciesAB061887 (Bacillus thuringiensis serovar alesti) [8] were used for the homology modelling. Homology modelling was done using the Swiss model workspace in the automatic modelling mode [http://swissmodel.expasy.org/] [1416]. The template used for homology modelling was 1V5CA. Protein data bank (PDB) homologs were determined for the sequences using the COFACTOR server [zhanglab.ccmb.med.umich.edu/COFACTOR/] [17]. RESULTS AND DISCUSSION Analysis of cellulase-chitosanase sequences from Bacillus species Chitosanolytic enzymes are gaining importance as they can produce low molecular weight compounds including chitooligomers with high yield and low pollution. They have numerous applications in http://bioinfo.aizeonpublishers.net/content/2014/1/bioinfo315-320.pdf biomedical, pharmaceutical, agricultural and food fields [18]. The following previously isolated cellulasechitosanases whose sequences were deposited in Genbank were selected for the in silico analysis. Chitosanase from Bacillus cereus H1, which showed CMC and glycol chitosan activity (Accession number Q849S1) [6] and soil bacterium Bacillus sp. strain KCTC 0377BP isolated by Choi et al. [7] (Accession number QALZ1). Lee et al. In 2007 screened several Bacillus thuringiensis subspecies which produced bifunctional cellulase-chitosanases (Accession numbers: AB061887, AB061888, AB061889, AB061891, AB061892, AB061893, AB061894, AB061895) [8]. Multiple sequence alignment and study of the GH-8 signature motif The retrieved protein sequences were aligned with Clustal Omega [www.ebi.ac.uk/Tools/msa/clustalo] [910] and the output of the multiple sequence alignment was obtained from Jalview [http://www.jalview.org/] [11]. The multiple sequence alignment output which contains the signature motif is shown in figure 1. The signature motif of the GH-8 family is a stretch of about 20 residues. The signature motif sequence has of two conserved aspartates. The first aspartate is believed to be a nucleophile in the catalytic mechanism [5]. The following consensus pattern was obtained: A-[ST]-D-[AG]-D-x(2)-[IM]-A-x-[SA]-[LIVM][LIVMG]-x-A-x(3)-[FW], where the first D is the active site residue. The signature motif was generated from Prosite [http://prosite.expasy.org/scanprosite/] [12] and the sequence logo was created. Sequence logo is a graphical representation of multiple sequence alignments. It has stacks of letters which are colour coded and represent amino acids at consecutive positions. Sequence logos are more precise than the consensus sequence. The total height of the logo is based on the degree of conservation of the sequence. The height of the amino acids in the stack pertains to the relative frequency of the amino acid at that position. The logos are read from the top of the stacks [20]. The colouring of the amino acids is based on their chemical properties. Polar amino acids have been coloured green (T- threonine, GGlycine, Y- Tyrosine, S- Serine), Basic amino acids are coloured blue (K- Lysine, H- Histidine), Acidic amino acids are coloured red (D- Aspartic acid) and hydrophobic amino acids (A- Alanine, L- Leucine and IIsoleucine) are coloured black [http://weblogo.berkeley.edu/info.html#color_sym]. Sequence logo was created using the online tool Weblogo [http://weblogo.berkeley.edu/logo.cgi] [13]. Sequence logo of the multiple sequence alignment is shown in figure 2. Homology modelling of cellulase-chitosanase sequences Homology modelling for the cellulase-chitosanase sequences was done in the automatic modelling mode 316 Niveditha Prakash and Shubha Gopal / Int J Comput Bioinfo In Silico Model. 2014, 3(1): 315-320 using the Swiss model server [http://swissmodel.expasy.org/] [14-16]. Homology modelling revealed that all the selected cellulasechitosanases sequences had the α6/α6 domain architecture which is typical of GH-8 members [5]. The homology models of the cellulase-chitosanase sequences with their signature motifs and catalytic domains are shown in figures 2, 3 and 4. Front view of the structures show the presence of the double barrel α6/α6 motif while the side views show the catalytic cleft and the N and C termini of the proteins. Signature motif of the proteins was seen in sequences 181-199. The first aspartate of the signature motif (Asp 183) was believed to be the active residue of the catalytic mechanism. The homology models were refined by using Modrefiner (http://zhanglab.ccmb.med.umich. edu/ModRefiner/) [21]. The quality of the structures that were modelled was verified with PROCHECK obtained from PDBsum. [http://www.ebi.ac.uk/ thornton-srv/databases/cgi-bin/pdbsum/] [21]. Figure 1. A part of the multiple sequence alignment showing the GH-8 signature motif. The signature motif is coloured purple. Figure 2. Sequence logo of the GH-8 signature motif. Sequence logo [http://weblogo.berkeley.edu/logo.cgi]. Signature motif is found in the position 181-199. (a) was generated from Weblogo (b) Table 1. Plot statistics for Bacillus cereus H1 chitosanase Residues in most favoured regions [A,B,L] Residues in additional allowed regions [a,b,l,p] Residues in generously allowed regions [~a,~b,~l,~p] Residues in disallowed regions 300 39 0 88.2% 11.5% 0.0% 1 0.3% Number of non-glycine and non-proline residues Number of end-residues (excl. Gly and Pro) Number of glycine residues (shown as triangles) Number of proline residues 340 100.0% Total number of residues 386 1 28 17 (c) Figure 3. Front view (a), side view (b), Ramachandran plot (c) and plot statistics for chitosanase from Bacillus cereus H1 http://bioinfo.aizeonpublishers.net/content/2014/1/bioinfo315-320.pdf 317 Niveditha Prakash and Shubha Gopal / Int J Comput Bioinfo In Silico Model. 2014, 3(1): 315-320 (d) (e) Table 2. Plot statistics for Bacillus sp. strain KCTC 0377BP Residues in most favoured regions [A,B,L] Residues in additional allowed regions [a,b,l,p] 305 89.7% 34 10% 0 1 0.0% 0.3% Number of non-glycine and non-proline residues Number of end-residues (excl. Gly and Pro) 340 1 100% Number of glycine residues (shown as triangles) Number of proline residues 28 17 Residues in generously allowed regions [~a,~b,~l,~p] Residues in disallowed regions Total number of residues 386 (f) Figure 4. Front view (d), side view (e), Ramachandran plot (f) and plot statistics for chitosanase Bacillus sp. strain KCTC 0377BP (g) (h) Table 3. Plot statistics for chitosanase, Bacillus thuringiensis serovar alesti Residues in most favoured regions [A,B,L] 304 89.4% Residues in additional allowed regions [a,b,l,p] 34 10.0% 1 0.3% 1 0.3% Number of non-glycine and non-proline residues Number of end-residues (excl. Gly and Pro) 340 1 100.0% Number of glycine residues (shown as triangles) Number of proline residues 28 17 Residues in generously allowed regions [~a,~b,~l,~p] Residues in disallowed regions Total number of residues 386 (i) Figure 5. Front view (g), side view (h), Ramachandran plot (i) and plot statistics for chitosanase Bacillus thuringiensis serovar alesti http://bioinfo.aizeonpublishers.net/content/2014/1/bioinfo315-320.pdf 318 Niveditha Prakash and Shubha Gopal / Int J Comput Bioinfo In Silico Model. 2014, 3(1): 315-320 Identification of structural homologs In order to detect the occurrence of proteins with structural similarity, the COFACTOR server was used [zhanglab.ccmb.med.umich.edu/COFACTOR/] [16]. The top 10 identified structural analogs from PDB were retrieved and tabulated as shown in table 4. All the structures share the common α6/α6 domain. This clearly shows that the cellulase-chitosanases from the selected Bacillus species are structurally related to cellulases, xylanases, endoglucanases, epimerases and terpenoid cyclases from different organisms. Table 4. Top 10 structural analogs of chitosanases from Bacillus cereus H1, Bacillus sp. strain KCTC 0377BP and Bacillus thuringiensis serovar alesti as identified from the COFACTOR server Sl. No. Chitosanase, Bacillus cereus H1 Chitosanase, Bacillus sp. strain KCTC 0377BP B.thuringiensis serovar cellulase-chitosanase structural analogs name of the enzyme structural analogs structural analogs Name of the enzyme 1 1v5cA Chitosanase 1l2aA Processive endocellulase CelF 1l2aA 2 1cemA Cellulase CelA 2afaB 3 2b4fA Endo-1,4-β-xylanase 3gt5A N-acylglucosamine (AGE) N-acylglucosamine (AGE) 3gt5A Processive endocellulase CelF N-acylglucosamine 2-epimerase (AGE) N-acylglucosamine 2-epimerase (AGE) name of the enzyme 2-epimerase 2afaB 2-epimerase alesti 4 2drrA Xylanase Y 3qxqA Endoglucanase 3qxqA Endoglucanase 5 4fusA Endoglucanase F 1wzzA Probable endoglucanase (cmcAX) 1wzzA 6 4el8A Endoglucanase F 1fp3B N-acylglucosamine (AGE) 2-epimerase 1fp3B Probable endoglucanase (cmcAX) N-acylglucosamine 2-epimerase (AGE) 7 2zblB N-acylglucosamine 2-epimerase (AGE) 3vw5A N-acylglucosamine (AGE) 2-epimerase 3vw5A N-acylglucosamine 2-epimerase (AGE) 8 2rgkC N-acylglucosamine 2-epimerase (AGE) 2zzrA Glycosyl hydrolase 2zzrA Glycosyl hydrolase 9 1l2aA 2fuzA Glycosyl hydrolase 2fuzA Glycosyl hydrolase 10 3gt5A Processive endocellulase CelF N-acylglucosamine 2-epimerase (AGE) 1w6jA Terpenoid cyclase 1w6kA Terpenoid cyclase CONCLUSION GH-8 is a diverse group of multifunctional enzymes having chitosanases, cellulases, xylanases, lichenases etc. Cellulase-chitosanases belonging to this family are gaining importance as they can produce low molecular weight chitooligomers which are commercially important for the pharmaceutical, agricultural and food industries. Bacillus species which are common soil inhabitants are known to produce cellulasechitosanases [3]. Several cellulase-chitosanases have been isolated so far, but their structural details are not clearly understood. The protein sequences of cellulasechitosanases were shown to have a signature motif of about 20 amino acids were the first aspartate is believed to be the nucleophile in the catalytic mechanism. All GH-8 proteins analysed here were found to have the α6/α6 double barrel structure. Significant structural similarity was seen among cellulases, xylanases, epimerses, endoglucanases and terpenoid cyclases from different organisms. This probably indicates the evolutionary relationship among different glycoside hydrolases having the α6/α6 double barrel catalytic core and the evolutionary stability of the α6/α6 motif among the glycoside hydrolases of several species [5]. An attempt has been made in this study to understand the structures of these beneficial http://bioinfo.aizeonpublishers.net/content/2014/1/bioinfo315-320.pdf enzymes and also establish their similarity with enzymes having related structures and function. REFERENCES 1. 2. 3. 4. 5. 6. 7. 8. 9. Mitsutomi M, Isono M, Uchiyama A, Nikaidou N, Ikegami T, Watanabe T (1998) Chitosanase activity of the enzyme previously reported as β-1,3-1,4-glucanase from Bacillus circulans WL-12. Biosci., Biotechnol., Biochem. 62: 2107-2114 Henrissat B and Davies G (1997) Structural and sequence-based classification of glycoside hydrolases. Curr. Opin. Struct. Biol. 7:637–644 Xia W, Liu P, Liu J (2008) Advance in chitosan hydrolysis by nonspecific cellulases. Bioresour. Technol. 99:6751-6762 Adachi W, Sakihama Y, Shimizu S et al. (2004) A Crystal structure of family GH-8 chitosanase with subclass II specificity from Bacillus sp. K17. J. Mol. Biol. 343:785-795 Alzari PM, Souchon H, Dominguez R (1996) The crystal structure of endoglucanase CelA, a family 8 glycosyl hydrolase from Clostridium thermocellum. Structure 4: 265-275 Jang HK, Yi JH, Kim JT et al. (2003) Purification, characterization and gene cloning of chitosanase from Bacillus cereus H-1. Korean J. Microb. Biotech. 31:216-223 Choi YJ, Kim EJ, Piao Z et al. (2004) Purification and characterization of chitosanase from Bacillus sp. strain KCTC 0377BP and its application for the production of chitosan oligosaccharides. Appl. Environ. Microbiol. 70: 4522–4531 Lee HS, Jang JS, Choi SK et al. (2007) Identification and expression of GH-8 family chitosanases from several Bacillus thuringiensis subspecies. FEMS Microbiol. Lett. 277:133–141 Sievers F, Wilm A, Dineen DG et al. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539 319 Niveditha Prakash and Shubha Gopal / Int J Comput Bioinfo In Silico Model. 2014, 3(1): 315-320 10. Goujon M, Mc William H, Li W et al. (2010) A new bioinformatics analysis tools framework at EMBL-EBI. Nucl. Acids Res. 38: W695-W699 11. Waterhouse AM, Procter JB, Martin DMA et al. (2009) Jalview Version 2 – a multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189-1191 12. De Castro E, Sigrist CJA, Gattiker A et al. (2006) ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucl. Acids Res. 34: W362-W365 13. Crooks GE, Hon G, Chandonia JM et al. (2004) Weblogo: A sequence logo generator. Genome Res. 14:1188-1190 14. Arnold K, Bordoli L, Kopp J et al. (2006) The SWISS-MODEL Workspace: A web-based environment for protein structure homology modelling. Bioinformatics 22: 195-201. 15. Kiefer F, Arnold K, Künzli M et al. (2009) The SWISS-MODEL Repository and associated resources. Nucl. Acids Res. 37: D387D392 16. Peitsch MC (1995) Protein modeling by E-mail. Nat. Biotechnol. 13: 658 - 660 17. Roy A, Yang J, Zhang Y (2012) COFACTOR: An accurate comparative algorithm for structure-based protein function annotation. Nucl. Acids Res. 40: W471-W477 18. Liu J and Xiaa JLW (2006) Purification and characterization of a bifunctional enzyme with chitosanase and cellulase activity from commercial cellulase. Biochem. Eng. J. 30:82-87 19. Schneider TD and Stephens RM (1990) Sequence Logos: A New Way to Display Consensus Sequences. Nucl. Acids Res. 18:60976100 20. Xu D and Zhang Y (2011) Improving the Physical Realism and Structural Accuracy of Protein Models by a Two-step Atomiclevel Energy Minimization. Biophys. J. 101: 2525-2534 21. Laskowski RA, MacArthur MW, Moss DS (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26: 283-291 © 2014; AIZEON Publishers; All Rights Reserved This is an Open Access article distributed under the terms of the Creative Commons Attribution License which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. ***** http://bioinfo.aizeonpublishers.net/content/2014/1/bioinfo315-320.pdf 320