* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Document
Photosynthetic reaction centre wikipedia , lookup
Magnesium transporter wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Fatty acid synthesis wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Fatty acid metabolism wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Western blot wikipedia , lookup
Point mutation wikipedia , lookup
Peptide synthesis wikipedia , lookup
Homology modeling wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Metalloprotein wikipedia , lookup
Calciseptine wikipedia , lookup
Genetic code wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Biosynthesis wikipedia , lookup
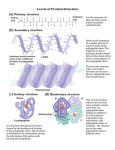
1 Incidentally, note the functional groups we have met so far: Hydroxyl Amine Amide Carboxyl Carbonyl Aldehyde Ketone Ester: Carboxylic acid ester Phosphoester And: Glycosidic bonds C=C double bonds (cis and trans) 2 PROTEINS Amino acids (the monomer of proteins) 3 At pH 7, ,most amino acids are zwitterions (charged but electrically neutral) 4 5 +OH- ( -H+) Net charge +H+ 50-50 charged-uncharged at ~ pH2.5 (=the pK) 50-50 charged-uncharged at ~ pH9 (=the pK) 6 Numbering (lettering) amino acids ε-amino group ε δ γ β Alpha-amino Alpha-carboxyl (attached to the α-carbon) Alpha-carbon Amino acid examples 7 Molecular weights 75 – 203 (MW) Glycine (gly) Side chain = H Smallest (75) One neg. charge β-carboxyl: -CH2-COOH Tryptophan (trp) 5+6 membered rings Hydrophobic, largest (203) Lysine (lys) One pos. charge ε-amino Alanine (ala) One carbon (methyl group) -CH3 Arginine (arg, guanidino group) One pos. charge -(NH-C (NH2)NH2)+, Aspartic acid (asp, aspartate) 8 Shown uncharged (as on exams) 9 10 11 Amino acids in 3 dimensions • Asymmetric carbon (4 different groups attached) • Stereoisomers • Rotate polarized light • Optical isomers • Non-superimposable • Mirror images • L and D forms From Purves text 12 Mannose 13 (Without showing the R-groups) The backbone is monotonous It is the side chains that provide the variety 14 “Polypeptides” vs. “proteins” • Polypeptide = amino acids connected in a linear chain (polymer) • Protein = a polypeptide or several associated polypeptides (discussed later) • Often used synonymously • Peptide (as opposed to polypeptide) is smaller, even 2 AAs (dipeptide) 15 The backbone is monotonous (Without showing the R-groups) It is the side chains that provide the variety 16 Proteins do most of the the jobs in the cell E.g., egg albumin, hemoglobin, keratin, estrogen receptor, immunoglobulins (antibodies), enzymes (e.g., beta-galactosidase) Each is a polymer or assemblage of polymers made up of amino acids Each particular protein polymer (polypeptide) has a unique sequence of amino acids Each molecule of a particular protein has the same sequence of amino acids. E.g., met-ala-leu-leu-arg-glu-leu-val- . . . . How is this sequence determined? 17 Primary (1o) Structure = the sequence of the amino acids in the polypeptide chain 18 Determining the sequence One way: enzyme: Carboxypeptidase: hydrolyzes the peptide bond , identify e.g., …. arg-leu-leu-val-gly-ala-gly-phe-trp-lys-glu-asp-ser …. arg-leu-leu-val-gly-ala-gly-phe-trp-lys-glu-asp …. arg-leu-leu-val-gly-ala-gly-phe-trp-lys-glu asp ser 19 AA mixture (-) (+) 20 A paper electrophoresis apparatus 21 Side view AAs applied at lower end 22 “Rf” 0.82 After stopping the paper chromatography and staining for the amino acids: 0.69 0.45 0.27 0.11 23 Paper chromatography apparatus 24 • Treatment of a polypeptide with trypsin • Trypsin is a proteolytic enzyme. • It catalyzes cleavage (hydrolysis) after lysine and arginine residues Polypeptide chain Sub-peptides The order of the subpeptides is unknown. The sequence is reconstructed by noting the overlap between differently produced subpeptides 25 Trypsin (lys, arg) (1) Chymotrypsin (trp, tyr, phe) (2) N C 26 Fingerprinting a protein: analysis of the sub-peptides (without breaking them down to their constituent amino acids) Application to sickle cell disease (Ingram, 1960’s) Hemoglobin protein Sub-peptides No further digestion to amino acids; left as sub-peptides 27 Oligopeptides behave as a composite of their constituent amino acids + H H H - Net charge = -1: moves toward the anode in paper electrophoreses Fairly hydrophobic (~5/6): expected to move moderately well in paper chromatography Nomenclature: ala-tyr-glu-pro-val-trp or AYEPVW or alanyl-tyrosyl-glutamyl-prolyl-valyl-tryptophan Hb 28 In fingerprinting, these spots contain peptides, not amino acids trypsin The mixture of all sub-peptides formed Less negatively charged, More hydrophobic Negatively charged ---valine--(sickle) Positively charged More hydrophobic More hydrophilic Negatively charged Positively charged Negatively charged Positively charged ---glutamate--(normal) 29 Every different polypeptide has a different primary structure (sequence). Every polypeptide will have different arrangement of spots after fingerprinting. 30 3-dimensional structure of proteins One given purified polypeptide • Molecule #1: N-met-leu-ala-asp-val-val-lys-.... • Molecule #2: N-met-leu-ala-asp-val-val-lys-... • Molecule #3: N-met-leu-ala-asp-val-val-lys-... • . Molecule #4: N-met-leu-ala-asp-val-val-lys-... etc 31 3-D structure of polypeptides Arrangement 1 at one moment in time? Arrangement 2 at one moment in time? Arrangement 3 at one moment in time? Arrangement 3 at one moment in time? [Rope models here] 32 33 34 Got this far 35 Primary structure itself results in some folding constraints: See bottom of handout 3-3 36 37 4 atoms in one plane 6 atoms in one plane 38 39 40 41 42 There’s still plenty of flexibility Secondary structure: the alpha helix 43 Amino acids shown simplified, without side chains and H’s. Alpha helix depictions 44 C = grays N = blue O = red Poly alanine Side chains = -CH3 (lighter gray) H’s not shown 45 Linus Pauling and a model of the alpha helix.1963 Secondary structure: H-bond AA residue 46 Beta sheet 47 48 Beta-sheets Anti-parallel Parallel 49 secondary structure (my definition): structure produced by regular repeated interactions between atoms of the backbone. Tertiary structure: The overall 3-D structure of a polypeptide. Neither Beta-sheets Alpha-helices These “ribbon” depictions do not show the side chains, only the backbone 50 Tertiary structure (overall 3-D) ionic hydrophobic H-bond cys Ionic-H combo covalent Van der Waals Weak bonds also occur throughout the polypeptide, between the amino acid side chains in regions of secondary structure as well as the looped regions 51 52 Disulfide bond formation Disulfide bond Sulfhydryl group R-CH2-SH cysteine + HS-CH2-R cysteine ½ O2 R-CH2-S-S-CH2-R + HOH cystine Two sulfhydryls have been oxidized (lost H’s) Oxygen has been reduced (gained H’s). Oxygen was the oxidizing agent. An oxidation-reduction reaction: Cysteines are getting oxidized (losing H atoms, with electron; NOT losing a proton, not like acids. Oxygen is getting reduced, gaining H-atoms and electrons Actually it’s the loss and gain of the electrons that constitutes oxidation and reduction, respectively. No catalyst usually needed. Overall 3-D structure of a polypeptide is tertiary structure 53 Stays intact in the jacuzzi at 37 deg C Usually does not require the strong covalent disulfide bond to maintain its 3-D structure Tuber model Protein structures are depicted in a variety of ways Backbone only Space-filing, With surface charge Ribbon Space-filling Blue = negative charge Red = positive 54 55 Information for proper exact folding (How does a polypeptide fold correctly?) Predicting protein 3-dimensional structure Determining protein 3-dimensional structure Where is the information for choosing the correct folded structure? Is it in the primary structure itself?