Editor(s): Laura Hoopes | http://www.nature.com/scitable/topic/gene
... differences in the genes each cell expresses. A cancer cell acts different from a normal cell for the same reason: It expresses different genes. (Using microarray analysis, scientists can use such differences to assist in diagnosis and selection of appropriate cancer treatment.) Interestingly, in eu ...
... differences in the genes each cell expresses. A cancer cell acts different from a normal cell for the same reason: It expresses different genes. (Using microarray analysis, scientists can use such differences to assist in diagnosis and selection of appropriate cancer treatment.) Interestingly, in eu ...
Plant Molecular Biology
... 1. These mutants show evidence of leaf development in darkness: they have expanded cotyledons, plastids that resemble chloroplasts, and chlorophyll protein genes turned on. 2. In the dark, these genes repress photomorphogenesis –related genes in all tissues. 3. In the light, they repress them only i ...
... 1. These mutants show evidence of leaf development in darkness: they have expanded cotyledons, plastids that resemble chloroplasts, and chlorophyll protein genes turned on. 2. In the dark, these genes repress photomorphogenesis –related genes in all tissues. 3. In the light, they repress them only i ...
Document
... • Sensitivity to DNAse reflects a relaxed chromatin structure that allows binding of transcription factors • The LCR regulates the entire gene cluster permitting it to be further regulated on a gene-by-gene basis ...
... • Sensitivity to DNAse reflects a relaxed chromatin structure that allows binding of transcription factors • The LCR regulates the entire gene cluster permitting it to be further regulated on a gene-by-gene basis ...
Gene regulation - Department of Plant Sciences
... • Transcriptional activator-like (TAL) effectors bind with plant promoters to express genes beneficial for the bacteria ...
... • Transcriptional activator-like (TAL) effectors bind with plant promoters to express genes beneficial for the bacteria ...
Ch. 19 – Eukaryotic Genomes
... Transcription initiation complex – resembles a prok promoter by being upstream Control elements – segments of noncoding DNA that regulate transcription by binding transcription factors Enhancers – help bend DNA for transcription factors, can be far from gene, even downstream Activators – help to pos ...
... Transcription initiation complex – resembles a prok promoter by being upstream Control elements – segments of noncoding DNA that regulate transcription by binding transcription factors Enhancers – help bend DNA for transcription factors, can be far from gene, even downstream Activators – help to pos ...
AP Biology Chapter 18, 19, 27 Study Guide Chapter 18: Regulation
... 3. What is differential gene expression? ...
... 3. What is differential gene expression? ...
Ch. 13.3 13.4 notes mutations
... Discovered in fruit flies but are found in all animals including humans ...
... Discovered in fruit flies but are found in all animals including humans ...
Chapter 12 DNA and RNA - Northwestern High School
... • Every cell can express different genes. – Pancreas secretes many digestive enzymes, amylase, that help break down starches. Expression of this genes allows it to function. Our marrow cells would not need to have this protein produced. – Morphogenesis (cell differentiation, cell specialization) ...
... • Every cell can express different genes. – Pancreas secretes many digestive enzymes, amylase, that help break down starches. Expression of this genes allows it to function. Our marrow cells would not need to have this protein produced. – Morphogenesis (cell differentiation, cell specialization) ...
reduce
... • A new method for discovering cis-regulatory elements • A new method for discovering cis-regulatory elements • A single genome-wide set of expression ratios, The upstream sequence for each gene, Outputs statistically significant motifs. Extract biologically meaningful information ...
... • A new method for discovering cis-regulatory elements • A new method for discovering cis-regulatory elements • A single genome-wide set of expression ratios, The upstream sequence for each gene, Outputs statistically significant motifs. Extract biologically meaningful information ...
Ch 15 Gudied Reading
... chromosomes. What is a plausible mechanism for the coordination of gene expression? ...
... chromosomes. What is a plausible mechanism for the coordination of gene expression? ...
A Gene Expression Experiment – Practical
... 1. Repeat the analysis of the liver and lung data set in the lecture 2. Look for sets of transcripts that have different patterns of expression between liver and lung. For example, you might look for genes which are expressed in both tissues but are not correlated, or look for genes expressed in one ...
... 1. Repeat the analysis of the liver and lung data set in the lecture 2. Look for sets of transcripts that have different patterns of expression between liver and lung. For example, you might look for genes which are expressed in both tissues but are not correlated, or look for genes expressed in one ...
LECT37 regul
... Q: Of this number how many are protein-encoding components? A: Roughly 1.5 percent Q: Have all of the genes been identified? A: No, we are not even close Q: What is left to do? A: Gene products, i.e., functional mRNAs and proteins, need to be identified, non-coding regulatory sequences need to be un ...
... Q: Of this number how many are protein-encoding components? A: Roughly 1.5 percent Q: Have all of the genes been identified? A: No, we are not even close Q: What is left to do? A: Gene products, i.e., functional mRNAs and proteins, need to be identified, non-coding regulatory sequences need to be un ...
Regulation of Gene Activity
... initiate/regulate transcription Posttranscriptional control: mRNA processing and how fast mRNA leaves the nucleus Translational control: when translation begins and how long it continues Posttranslational control: after protein synthesis, polypeptide may have to undergo additional changes before it ...
... initiate/regulate transcription Posttranscriptional control: mRNA processing and how fast mRNA leaves the nucleus Translational control: when translation begins and how long it continues Posttranslational control: after protein synthesis, polypeptide may have to undergo additional changes before it ...
Regulatory genes
... 3 parts to an operon 1. Operator – controls access of RNA polymerase to the promoter 2. Promoter – where RNA polymerase attaches to begin transcription of genes 3. Genes – code for expression of proteins related to one particular function (e.g. breaking down galactosidase) ...
... 3 parts to an operon 1. Operator – controls access of RNA polymerase to the promoter 2. Promoter – where RNA polymerase attaches to begin transcription of genes 3. Genes – code for expression of proteins related to one particular function (e.g. breaking down galactosidase) ...
Biology 303 EXAM II 3/14/00 NAME
... A. a process that only bacteria perform since they contain no nucleus. B. a process that is exclusively associated with transcription by RNA polymerase III in eukaryotes. C. alteration in chromatin structure to facilitate loading and translation by ribosomes and, thus, enhance gene expression. D. al ...
... A. a process that only bacteria perform since they contain no nucleus. B. a process that is exclusively associated with transcription by RNA polymerase III in eukaryotes. C. alteration in chromatin structure to facilitate loading and translation by ribosomes and, thus, enhance gene expression. D. al ...
Genetic Organization and Control
... 3. What did she create using crossover frequencies? 4. What does Ds stand for? Ac? 5. Which locus can insert into the colored gene and disrupt it? 6. What color will corn be if the colored gene is disrupted? 7. What is the term used for the gene “jumping” from one spot on the chromosome to another? ...
... 3. What did she create using crossover frequencies? 4. What does Ds stand for? Ac? 5. Which locus can insert into the colored gene and disrupt it? 6. What color will corn be if the colored gene is disrupted? 7. What is the term used for the gene “jumping” from one spot on the chromosome to another? ...
Jeopardy
... 100 What does the “mi” in miRNA represent? 100 bonus: siRNA? 200 What protein breaks up RNA transcripts from miRNA-encoding genes? 300 What are the two results of miRNA binding to a target mRNA? 400 Describe the process of ubiquination. 400 bonus: ubiquination comes after which step in protein produ ...
... 100 What does the “mi” in miRNA represent? 100 bonus: siRNA? 200 What protein breaks up RNA transcripts from miRNA-encoding genes? 300 What are the two results of miRNA binding to a target mRNA? 400 Describe the process of ubiquination. 400 bonus: ubiquination comes after which step in protein produ ...
Lecture#7 - Eukaryote gene structure and regulation.
... type of mature mRNA - > more than one type of protein product The same primary transcript can be processed to yield different mRNAs, which encode different proteins. (Another means of gene regulation - post-transcriptional level) ...
... type of mature mRNA - > more than one type of protein product The same primary transcript can be processed to yield different mRNAs, which encode different proteins. (Another means of gene regulation - post-transcriptional level) ...
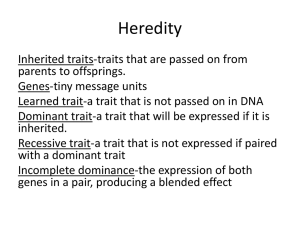
Heredity
... parents to offsprings. Genes-tiny message units Learned trait-a trait that is not passed on in DNA Dominant trait-a trait that will be expressed if it is inherited. Recessive trait-a trait that is not expressed if paired with a dominant trait Incomplete dominance-the expression of both genes in a pa ...
... parents to offsprings. Genes-tiny message units Learned trait-a trait that is not passed on in DNA Dominant trait-a trait that will be expressed if it is inherited. Recessive trait-a trait that is not expressed if paired with a dominant trait Incomplete dominance-the expression of both genes in a pa ...