Other Plasmid Maps Feature list descriptions
... H3K9 methylation and chromatin condensation. (mcb.asm.org/content/23/14/4753.short) ...
... H3K9 methylation and chromatin condensation. (mcb.asm.org/content/23/14/4753.short) ...
8.6 Gene Expression and Regulation
... • An operator is a part of DNA that turns a gene “on” or ”off.” • An operon includes a promoter, an operator, and one or more structural genes that code for all the proteins needed to do a job. – Operons are most common in prokaryotes. – The lac operon was one of the first examples of gene regulatio ...
... • An operator is a part of DNA that turns a gene “on” or ”off.” • An operon includes a promoter, an operator, and one or more structural genes that code for all the proteins needed to do a job. – Operons are most common in prokaryotes. – The lac operon was one of the first examples of gene regulatio ...
258927_Fx_DNA-RNA
... 15. In this example, what occurs during translation? 16. What must be done to this string of amino acids in order to turn it into a functional protein? 17. The rest of this process isn’t really about transcription or translation, but rather about enzymatic activity and is thus beyond the scope of ou ...
... 15. In this example, what occurs during translation? 16. What must be done to this string of amino acids in order to turn it into a functional protein? 17. The rest of this process isn’t really about transcription or translation, but rather about enzymatic activity and is thus beyond the scope of ou ...
Name:
... 15. In this example, what occurs during translation? 16. What must be done to this string of amino acids in order to turn it into a functional protein? 17. The rest of this process isn’t really about transcription or translation, but rather about enzymatic activity and is thus beyond the scope of ou ...
... 15. In this example, what occurs during translation? 16. What must be done to this string of amino acids in order to turn it into a functional protein? 17. The rest of this process isn’t really about transcription or translation, but rather about enzymatic activity and is thus beyond the scope of ou ...
Chapter 1 Study Questions
... 4. Compare the chemical structure of a small non-polar amino acid (such as alanine) to one with a bulky hydrocarbon side chain (such as isoleucine). What kind of chemical interactions are non-polar side chains involved in? 5. Which amino acids contain sulfur? Which contain hydroxyl (-OH) groups? Whi ...
... 4. Compare the chemical structure of a small non-polar amino acid (such as alanine) to one with a bulky hydrocarbon side chain (such as isoleucine). What kind of chemical interactions are non-polar side chains involved in? 5. Which amino acids contain sulfur? Which contain hydroxyl (-OH) groups? Whi ...
MS Word file
... Terminator-halts transcription and releases RNA molecule Initiation The substrate for transcription: Ribonucleoside triphosphates – rNTPs added to the 3′ end of the RNA molecule rGTP, rCTP, rATP, and rUTP Initiation The transcription apparatus: Bacterial RNA polymerase: five subunits made up of the ...
... Terminator-halts transcription and releases RNA molecule Initiation The substrate for transcription: Ribonucleoside triphosphates – rNTPs added to the 3′ end of the RNA molecule rGTP, rCTP, rATP, and rUTP Initiation The transcription apparatus: Bacterial RNA polymerase: five subunits made up of the ...
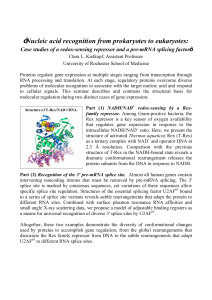
Nucleic acid recognition from prokaryotes to eukaryotes: Case
... Part (2) Recognition of the 3' pre-mRNA splice site. Almost all human genes contain intervening noncoding introns that must be removed by pre-mRNA splicing. The 3' splice site is marked by consensus sequences, yet variations of these sequences allow specific splice site regulation. Structures of the ...
... Part (2) Recognition of the 3' pre-mRNA splice site. Almost all human genes contain intervening noncoding introns that must be removed by pre-mRNA splicing. The 3' splice site is marked by consensus sequences, yet variations of these sequences allow specific splice site regulation. Structures of the ...
Transcription and Translation
... different amounts of molecules) I.e. difference in antibodies (some get sick more often or from different things) ...
... different amounts of molecules) I.e. difference in antibodies (some get sick more often or from different things) ...
Previously in Bio308
... How would a neuropeptide get made (in general terms)? What are the basic parts of DNA, RNA, and proteins? What is the difference between hnRNA, mRNA and tRNA? ...
... How would a neuropeptide get made (in general terms)? What are the basic parts of DNA, RNA, and proteins? What is the difference between hnRNA, mRNA and tRNA? ...
Control of Eukaryotic Gene Expression (Learning Objectives)
... 2. Explain the role of chemical modifications: methylation of DNA and acetylation of histones in control of gene expression. Define the term epigenetics. 3. Identify the main mechanism for turning on gene expression. Explain why control of gene expression in eukaryotic cells is like a “dimmer switch ...
... 2. Explain the role of chemical modifications: methylation of DNA and acetylation of histones in control of gene expression. Define the term epigenetics. 3. Identify the main mechanism for turning on gene expression. Explain why control of gene expression in eukaryotic cells is like a “dimmer switch ...
10chap19guidedreadingVideo
... 5. IF cells carry all of the genetic differences, why then are cells so unique – what is responsible for this? 6. In the diagram below – highlight all of the potential locations for gene expression regulation in eukaryotic cells. How does this compare with prokaryotic cells? ...
... 5. IF cells carry all of the genetic differences, why then are cells so unique – what is responsible for this? 6. In the diagram below – highlight all of the potential locations for gene expression regulation in eukaryotic cells. How does this compare with prokaryotic cells? ...
chapter13
... - A gene having one UPE is generally weakly translated, whereas one containing fire or six UPEs is usually actively translated. ...
... - A gene having one UPE is generally weakly translated, whereas one containing fire or six UPEs is usually actively translated. ...
Chapter 15 - jl041.k12.sd.us
... added to the 5’ end and a poly-A tail added to the 3’ end. If these tails/caps are long, it will take the enzymes in the cytoplasm a greater amount of time to digest the coding region of the transcript. ...
... added to the 5’ end and a poly-A tail added to the 3’ end. If these tails/caps are long, it will take the enzymes in the cytoplasm a greater amount of time to digest the coding region of the transcript. ...
Trnascription in eucaryotes
... control of initiation and control of gene transcription in prokaryotes and eukaryotes • Unlike in prokaryotes RNA polymerase does not recognize sites on the DNA itself but binds because a large number of other proteins bind and recruit the polymerase. • A bacterium has about 4000 genes but a mammal ...
... control of initiation and control of gene transcription in prokaryotes and eukaryotes • Unlike in prokaryotes RNA polymerase does not recognize sites on the DNA itself but binds because a large number of other proteins bind and recruit the polymerase. • A bacterium has about 4000 genes but a mammal ...
What is trans-acting factor?
... protein. Example: the activator Gal4 is controlled by the masking Gal80). Some masking proteins not only block the activating region of an activator but also recruit a deacetylase enzyme to repress the target genes. Example: Rb represses the function of the mammalian transcription activator E2F in t ...
... protein. Example: the activator Gal4 is controlled by the masking Gal80). Some masking proteins not only block the activating region of an activator but also recruit a deacetylase enzyme to repress the target genes. Example: Rb represses the function of the mammalian transcription activator E2F in t ...
Nucleic Acids - faculty at Chemeketa
... What will be the composition of the DNA strand complementary to –AGCCA– ? a. b. c. d. ...
... What will be the composition of the DNA strand complementary to –AGCCA– ? a. b. c. d. ...
4.3 DNA Control Mechanisms
... DNA Control Mechanisms in all cells (Remember, these are ways to control Gene Expression.) A. Transposons “Jumping Genes” (These DNA segments act as “Blockers” to transcription.) 1. Barbara McClintock discovered this control mechanism in the1940’s. She worked with Maize. She won a Nobel Prize for th ...
... DNA Control Mechanisms in all cells (Remember, these are ways to control Gene Expression.) A. Transposons “Jumping Genes” (These DNA segments act as “Blockers” to transcription.) 1. Barbara McClintock discovered this control mechanism in the1940’s. She worked with Maize. She won a Nobel Prize for th ...
PPT File
... This then binds to a proteasome, a protein complex which contains enzymes. The polyubiquitin is removed and ATP is used to unfold the protein. It is digested into peptides and amino acids. Cyclins in the cell cycle are regulated in this way. ...
... This then binds to a proteasome, a protein complex which contains enzymes. The polyubiquitin is removed and ATP is used to unfold the protein. It is digested into peptides and amino acids. Cyclins in the cell cycle are regulated in this way. ...
Gene Regulation
... GENE REGULATION Where does regulation occur? At what step? Most regulation occurs at the DNA to RNA step—transcription! Why? Conserves the most energy! ...
... GENE REGULATION Where does regulation occur? At what step? Most regulation occurs at the DNA to RNA step—transcription! Why? Conserves the most energy! ...
Document
... Protein degradation in eukaryotes requires a protein co-factor called ubiquitin. Ubiquitin binds to proteins and identifies them for degradation by proteolytic enzymes. ...
... Protein degradation in eukaryotes requires a protein co-factor called ubiquitin. Ubiquitin binds to proteins and identifies them for degradation by proteolytic enzymes. ...
Transcription
... • Enzymes attach to DNA at the gene’s location and unzip only where that gene is on the DNA. – DNA A T C G ...
... • Enzymes attach to DNA at the gene’s location and unzip only where that gene is on the DNA. – DNA A T C G ...
Gene Regulation
... – short segments of RNA (21-28 bases) • bind to mRNA • create sections of double-stranded mRNA • “death” tag for mRNA – triggers degradation of mRNA ...
... – short segments of RNA (21-28 bases) • bind to mRNA • create sections of double-stranded mRNA • “death” tag for mRNA – triggers degradation of mRNA ...
Document
... 1. Every step in transcription initiation can be regulated to increase or decrease the number of successful initiations per time. 2. In E. coli, transcription initiation is controlled primarily by alternative factors and by a large variety of other sequence-specific DNA-binding proteins. 3. G=RTl ...
... 1. Every step in transcription initiation can be regulated to increase or decrease the number of successful initiations per time. 2. In E. coli, transcription initiation is controlled primarily by alternative factors and by a large variety of other sequence-specific DNA-binding proteins. 3. G=RTl ...