HomeworkCh7
... c. What is a promotor? d. What are the three main phases of RNA synthesis? e. Can more than one copy of the gene be copied at the same time? 6. Translation a. What is translation? Why do you think it’s called that? b. How many different codons are possible for providing a three nucleotide code for t ...
... c. What is a promotor? d. What are the three main phases of RNA synthesis? e. Can more than one copy of the gene be copied at the same time? 6. Translation a. What is translation? Why do you think it’s called that? b. How many different codons are possible for providing a three nucleotide code for t ...
Site-specific recombination mechanisms exploit DNA
... bacteriophage (Mu) changes its host range through expression of different tail fibers by changing the orientation of a specific DNA segment, the G segment, in its genome1. The phage-encoded Gin recombinase protein specifically recombined the G segment located between short inverted DNA sequences, bu ...
... bacteriophage (Mu) changes its host range through expression of different tail fibers by changing the orientation of a specific DNA segment, the G segment, in its genome1. The phage-encoded Gin recombinase protein specifically recombined the G segment located between short inverted DNA sequences, bu ...
DNA Timeline Assignment
... It took him eight years and more than 10,000 pea plants to discover the laws of inheritance. _____________________________________________ ...
... It took him eight years and more than 10,000 pea plants to discover the laws of inheritance. _____________________________________________ ...
: Determining DNA sequences
... which has a coloured florescent marker attached. In has the added property of terminating the elongation if chosen instead of dATP • During the process all possible lengths of chain are produced. • Lengths are separated based on weight and analysed to give • The complementary sequence of the templat ...
... which has a coloured florescent marker attached. In has the added property of terminating the elongation if chosen instead of dATP • During the process all possible lengths of chain are produced. • Lengths are separated based on weight and analysed to give • The complementary sequence of the templat ...
Genes, Chromosomes, and DNA
... All organisms are made of one or more cells With few exceptions, all cells contain DNA All organisms have DNA ...
... All organisms are made of one or more cells With few exceptions, all cells contain DNA All organisms have DNA ...
DNA cr.eu updated plg latest
... with approximately 147 base pairs of DNA wound around them; in euchromatin, this wrapping is loose so that the raw DNA may be accessed. • Each core histone possesses a `tail' structure, which can vary in several ways; it is thought that these variations act as "master control switches," which determ ...
... with approximately 147 base pairs of DNA wound around them; in euchromatin, this wrapping is loose so that the raw DNA may be accessed. • Each core histone possesses a `tail' structure, which can vary in several ways; it is thought that these variations act as "master control switches," which determ ...
7529 DNA Sequencing - ACM
... Krusty Krab out of business. So, SpongeBob and his co-workers decided to switch to a brand new job. Their new startup is Krusty-Royan, a biological research institute whose main focus is on DNA sequencing. Their first customer is Sandy, the squirrel scientist, who has found the corpse of an alien fr ...
... Krusty Krab out of business. So, SpongeBob and his co-workers decided to switch to a brand new job. Their new startup is Krusty-Royan, a biological research institute whose main focus is on DNA sequencing. Their first customer is Sandy, the squirrel scientist, who has found the corpse of an alien fr ...
DNA REVIEW SHEET (answer in COMPLETE sentences on another
... Review Frederick Griffith’s experiment (1928) in detail. Why did S cells maintain ability to synthesize capsules while R cells could not? What does the term transformation mean in terms of DNA? Describe/diagram Avery et. al (1944) experiment. Describe and diagram the experiment performed by Alfred H ...
... Review Frederick Griffith’s experiment (1928) in detail. Why did S cells maintain ability to synthesize capsules while R cells could not? What does the term transformation mean in terms of DNA? Describe/diagram Avery et. al (1944) experiment. Describe and diagram the experiment performed by Alfred H ...
Chromosomal Microarray (CGH+SNP)
... chromosome analysis, as well as large imbalances detectable by routine chromosome analysis. A special slide (chip) is used that contains thousands of spots neatly arranged in a checkerboard pattern known as “microarray.” Each spot contains DNA segments (DNA probes) that are specific to certain genom ...
... chromosome analysis, as well as large imbalances detectable by routine chromosome analysis. A special slide (chip) is used that contains thousands of spots neatly arranged in a checkerboard pattern known as “microarray.” Each spot contains DNA segments (DNA probes) that are specific to certain genom ...
MCB Lecture 1 – Molecular Diagnostics
... What is the typical size of fragments that PCR can amplify? o >1kb How many cycles must you perform via PCR before you get the first exact sample that you want to amplify? o 4 Cycles If you have a single base difference in sequence that does not affect a restriction site, how do you detect it? o Use ...
... What is the typical size of fragments that PCR can amplify? o >1kb How many cycles must you perform via PCR before you get the first exact sample that you want to amplify? o 4 Cycles If you have a single base difference in sequence that does not affect a restriction site, how do you detect it? o Use ...
Chapter 10 Study Guide Know the definitions for: Cross
... Be able to draw and label the structure of a nucleotide and DNA ladder: Backbone or sides of the DNA ladder composed of _?_ & _?_ Rungs of DNA ladder composed of _?_ _?_ Nitrogen bases of DNAPurines (double-ring structure) consist of _?_ (G) & _?_ (A) Pyrimidines (single-ring structure) consist of _ ...
... Be able to draw and label the structure of a nucleotide and DNA ladder: Backbone or sides of the DNA ladder composed of _?_ & _?_ Rungs of DNA ladder composed of _?_ _?_ Nitrogen bases of DNAPurines (double-ring structure) consist of _?_ (G) & _?_ (A) Pyrimidines (single-ring structure) consist of _ ...
Genetic Engineering
... organism to another: Restriction enzymes were used naturally to cut out viral DNA from their own DNA and destroy it 1. Cut the DNA containing the gene of interest (GOI) away from the genes surrounding it ...
... organism to another: Restriction enzymes were used naturally to cut out viral DNA from their own DNA and destroy it 1. Cut the DNA containing the gene of interest (GOI) away from the genes surrounding it ...
Lecture 8 (2/15/10) "DNA Forensics, Cancer, and Sequencing"
... genetically susceptible to baldness. He was a palaeoEskimo, and by comparing his genome to other living people, they deduced that he was member of the Arctic Saqqaq, the first known culture to settle in Greenland whose ancestors had trekked from Siberia around the Arctic circle in pursuit of game. C ...
... genetically susceptible to baldness. He was a palaeoEskimo, and by comparing his genome to other living people, they deduced that he was member of the Arctic Saqqaq, the first known culture to settle in Greenland whose ancestors had trekked from Siberia around the Arctic circle in pursuit of game. C ...
High-Resolution Array-Based Comparative Genomic Hybridization
... Array-based CGH (aCGH) is a new method that weds traditional CGH and microarray technology. In aCGH, tumor and normal genomic DNAs are differentially fluorescently labeled and co-hybridized onto an array containing mapped DNA sequences, providing measurements of tumor copy number changes at high res ...
... Array-based CGH (aCGH) is a new method that weds traditional CGH and microarray technology. In aCGH, tumor and normal genomic DNAs are differentially fluorescently labeled and co-hybridized onto an array containing mapped DNA sequences, providing measurements of tumor copy number changes at high res ...
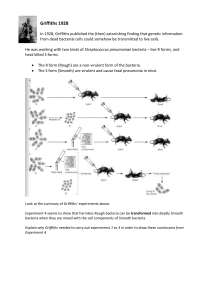
DNA experiments exercise
... bacteria when they are mixed with the cell components of Smooth bacteria. Explain why Griffiths needed to carry out experiments 1 to 3 in order to draw these conclusions from Experiment 4. ...
... bacteria when they are mixed with the cell components of Smooth bacteria. Explain why Griffiths needed to carry out experiments 1 to 3 in order to draw these conclusions from Experiment 4. ...
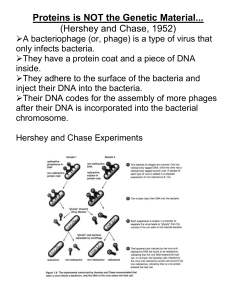
Hershey and Chase`s Experiment
... They adhere to the surface of the bacteria and inject their DNA into the bacteria. Their DNA codes for the assembly of more phages after their DNA is incorporated into the bacterial chromosome. Hershey and Chase Experiments ...
... They adhere to the surface of the bacteria and inject their DNA into the bacteria. Their DNA codes for the assembly of more phages after their DNA is incorporated into the bacterial chromosome. Hershey and Chase Experiments ...
DNA Fingerprinting Notes - Hicksville Public Schools
... ------------------------------------------------------------------------------------------------------------------------------1. Base your answer to the question on the diagram below and on your knowledge of biology. The diagram shows the results of a technique used to analyze DNA. This laboratory t ...
... ------------------------------------------------------------------------------------------------------------------------------1. Base your answer to the question on the diagram below and on your knowledge of biology. The diagram shows the results of a technique used to analyze DNA. This laboratory t ...
WEEK 1 PROBLEMS Problems From Chapter 1
... 1.1 In the early years of the twentieth century, why did many biologists and biochemists believe that proteins were probably the genetic material? 1.2 When the base composition of a DNA sample from Micrococcus luteus was determined, 37.5 percent of the bases were found to be cytosine. The DNA of thi ...
... 1.1 In the early years of the twentieth century, why did many biologists and biochemists believe that proteins were probably the genetic material? 1.2 When the base composition of a DNA sample from Micrococcus luteus was determined, 37.5 percent of the bases were found to be cytosine. The DNA of thi ...
Name - EdWeb
... 1. What is DNA? __________________________________________________________________ ______________________________________________________________________________ 2. What does DNA stand for? ________________________________________________________ 3. Why is DNA called a blueprint? ___________________ ...
... 1. What is DNA? __________________________________________________________________ ______________________________________________________________________________ 2. What does DNA stand for? ________________________________________________________ 3. Why is DNA called a blueprint? ___________________ ...
Presentación de PowerPoint
... further histone holding these together; Do not allow histone wrapped around DNA. Most of the DNA of a human cell is contained in the nucleus. Distinguish between unique and highly repetitive sequences in nuclear DNA. ...
... further histone holding these together; Do not allow histone wrapped around DNA. Most of the DNA of a human cell is contained in the nucleus. Distinguish between unique and highly repetitive sequences in nuclear DNA. ...
Comparative genomic hybridization

Comparative genomic hybridization is a molecular cytogenetic method for analysing copy number variations (CNVs) relative to ploidy level in the DNA of a test sample compared to a reference sample, without the need for culturing cells. The aim of this technique is to quickly and efficiently compare two genomic DNA samples arising from two sources, which are most often closely related, because it is suspected that they contain differences in terms of either gains or losses of either whole chromosomes or subchromosomal regions (a portion of a whole chromosome). This technique was originally developed for the evaluation of the differences between the chromosomal complements of solid tumor and normal tissue, and has an improved resoIution of 5-10 megabases compared to the more traditional cytogenetic analysis techniques of giemsa banding and fluorescence in situ hybridization (FISH) which are limited by the resolution of the microscope utilized.This is achieved through the use of competitive fluorescence in situ hybridization. In short, this involves the isolation of DNA from the two sources to be compared, most commonly a test and reference source, independent labelling of each DNA sample with a different fluorophores (fluorescent molecules) of different colours (usually red and green), denaturation of the DNA so that it is single stranded, and the hybridization of the two resultant samples in a 1:1 ratio to a normal metaphase spread of chromosomes, to which the labelled DNA samples will bind at their locus of origin. Using a fluorescence microscope and computer software, the differentially coloured fluorescent signals are then compared along the length of each chromosome for identification of chromosomal differences between the two sources. A higher intensity of the test sample colour in a specific region of a chromosome indicates the gain of material of that region in the corresponding source sample, while a higher intensity of the reference sample colour indicates the loss of material in the test sample in that specific region. A neutral colour (yellow when the fluorophore labels are red and green) indicates no difference between the two samples in that location.CGH is only able to detect unbalanced chromosomal abnormalities. This is because balanced chromosomal abnormalities such as reciprocal translocations, inversions or ring chromosomes do not affect copy number, which is what is detected by CGH technologies. CGH does, however, allow for the exploration of all 46 human chromosomes in single test and the discovery of deletions and duplications, even on the microscopic scale which may lead to the identification of candidate genes to be further explored by other cytological techniques.Through the use of DNA microarrays in conjunction with CGH techniques, the more specific form of array CGH (aCGH) has been developed, allowing for a locus-by-locus measure of CNV with increased resolution as low as 100 kilobases. This improved technique allows for the aetiology of known and unknown conditions to be discovered.