Chapter 15: Genetic Engineering
... DNA that have little/no function but that vary widely from one individual to another Use REs to cut DNA into fragments, electrophoresis to separate fragments ...
... DNA that have little/no function but that vary widely from one individual to another Use REs to cut DNA into fragments, electrophoresis to separate fragments ...
DNA - eduBuzz.org
... This characteristic number is known as the chromosome complement and varies from species to species. Each individuals DNA is unique. Genes are passed on from parent to offspring. Structure of chromosomes Chromosomes are made from tightly coiled molecules of a chemical substance called DNA. DNA All t ...
... This characteristic number is known as the chromosome complement and varies from species to species. Each individuals DNA is unique. Genes are passed on from parent to offspring. Structure of chromosomes Chromosomes are made from tightly coiled molecules of a chemical substance called DNA. DNA All t ...
Molecular characterization of individual DNA double strand breaks
... DNA double strand breaks (DSBs) are deadly lesions that can lead to genetic defects and cell apoptosis1. Techniques to directly image DSBs in isolated DNA include scanning electron microscopy2, Atomic Force Microscopy (AFM) and single molecule fluorescence microscopy3. While these techniques can be ...
... DNA double strand breaks (DSBs) are deadly lesions that can lead to genetic defects and cell apoptosis1. Techniques to directly image DSBs in isolated DNA include scanning electron microscopy2, Atomic Force Microscopy (AFM) and single molecule fluorescence microscopy3. While these techniques can be ...
Manipulating DNA
... • These cells could be used to generate new organs or cell clusters to treat patients with failing organs or degenerative diseases ...
... • These cells could be used to generate new organs or cell clusters to treat patients with failing organs or degenerative diseases ...
ch 20 study guide: dna technology
... Electro - = electricity (electroporation: a technique to introduce recombinant DNA into cells by applying a breif electrical pulse to a solution containing cells) Poly - = many; morph - = form (Single nucleotide polymorphisms: one-base-pair variations in the ...
... Electro - = electricity (electroporation: a technique to introduce recombinant DNA into cells by applying a breif electrical pulse to a solution containing cells) Poly - = many; morph - = form (Single nucleotide polymorphisms: one-base-pair variations in the ...
Factors Associated with Childhood Tumours (FACT) study
... Ideally 2x 9ml bottles, but we will include patients from whom less is available; please send samples from both parents if available, but we will include patients without parental samples. PLEASE SEND SAMPLES AND FORMS BY FIRST-CLASS POST TO: (PRE-PAID ADDRESS LABELS ARE AVAILABLE) ...
... Ideally 2x 9ml bottles, but we will include patients from whom less is available; please send samples from both parents if available, but we will include patients without parental samples. PLEASE SEND SAMPLES AND FORMS BY FIRST-CLASS POST TO: (PRE-PAID ADDRESS LABELS ARE AVAILABLE) ...
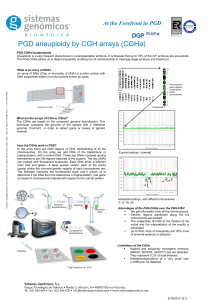
At the Forefront in PGD
... rearrangement (translocation or inversion) have an increased risk of generating abnormal embryos as a result of segregation of the balanced abnormality. This causes, recurrent abortions and, in many cases, infertility. PGD using FISH techniques allows detect altered embryos (unbalanced) for a specif ...
... rearrangement (translocation or inversion) have an increased risk of generating abnormal embryos as a result of segregation of the balanced abnormality. This causes, recurrent abortions and, in many cases, infertility. PGD using FISH techniques allows detect altered embryos (unbalanced) for a specif ...
ChIP-seq
... Use PCR to amplify specific DNA sequences to see if they were precipitated with the antibody. ...
... Use PCR to amplify specific DNA sequences to see if they were precipitated with the antibody. ...
EXAM 2
... True/False (1 point each) 20. ___T___ Satellite DNA is highly repetitive 21. ___T___ The more repetitive DNA included in a genome, the more quickly it will reanneal after being denatured. 22. ___T___ For most diploid eukaryotic organisms, sexual reproduction is the only mechanism resulting in new me ...
... True/False (1 point each) 20. ___T___ Satellite DNA is highly repetitive 21. ___T___ The more repetitive DNA included in a genome, the more quickly it will reanneal after being denatured. 22. ___T___ For most diploid eukaryotic organisms, sexual reproduction is the only mechanism resulting in new me ...
DNA
... *is passed from one generation to the next in chromosomes. *looks like a ladder, twisted around itself, called a double helix DNA Timeline Facts… Early 1950’s o 1st picture of DNA taken by Rosalind Franklin using an X-ray machine. ...
... *is passed from one generation to the next in chromosomes. *looks like a ladder, twisted around itself, called a double helix DNA Timeline Facts… Early 1950’s o 1st picture of DNA taken by Rosalind Franklin using an X-ray machine. ...
DNA History Function Structure
... always equal to the amount of T. – The amount of C is always equal to the amount of G. – What can be inferred? • A goes with T • G goes with C ...
... always equal to the amount of T. – The amount of C is always equal to the amount of G. – What can be inferred? • A goes with T • G goes with C ...
which together form the gene "stories" NOTE
... 1) the DNA strand will 'unzip' as the chemical bonds are broken between each of the nitrogen bases ...
... 1) the DNA strand will 'unzip' as the chemical bonds are broken between each of the nitrogen bases ...
DNA Notes - Firelands Local Schools
... SYNTHESIS. – DNA IS A SELF-REPLICATING MOLECULE WHICH GETS PASSED ON FROM ONE GENERATION TO THE NEXT. ...
... SYNTHESIS. – DNA IS A SELF-REPLICATING MOLECULE WHICH GETS PASSED ON FROM ONE GENERATION TO THE NEXT. ...
Comparative genomic hybridization

Comparative genomic hybridization is a molecular cytogenetic method for analysing copy number variations (CNVs) relative to ploidy level in the DNA of a test sample compared to a reference sample, without the need for culturing cells. The aim of this technique is to quickly and efficiently compare two genomic DNA samples arising from two sources, which are most often closely related, because it is suspected that they contain differences in terms of either gains or losses of either whole chromosomes or subchromosomal regions (a portion of a whole chromosome). This technique was originally developed for the evaluation of the differences between the chromosomal complements of solid tumor and normal tissue, and has an improved resoIution of 5-10 megabases compared to the more traditional cytogenetic analysis techniques of giemsa banding and fluorescence in situ hybridization (FISH) which are limited by the resolution of the microscope utilized.This is achieved through the use of competitive fluorescence in situ hybridization. In short, this involves the isolation of DNA from the two sources to be compared, most commonly a test and reference source, independent labelling of each DNA sample with a different fluorophores (fluorescent molecules) of different colours (usually red and green), denaturation of the DNA so that it is single stranded, and the hybridization of the two resultant samples in a 1:1 ratio to a normal metaphase spread of chromosomes, to which the labelled DNA samples will bind at their locus of origin. Using a fluorescence microscope and computer software, the differentially coloured fluorescent signals are then compared along the length of each chromosome for identification of chromosomal differences between the two sources. A higher intensity of the test sample colour in a specific region of a chromosome indicates the gain of material of that region in the corresponding source sample, while a higher intensity of the reference sample colour indicates the loss of material in the test sample in that specific region. A neutral colour (yellow when the fluorophore labels are red and green) indicates no difference between the two samples in that location.CGH is only able to detect unbalanced chromosomal abnormalities. This is because balanced chromosomal abnormalities such as reciprocal translocations, inversions or ring chromosomes do not affect copy number, which is what is detected by CGH technologies. CGH does, however, allow for the exploration of all 46 human chromosomes in single test and the discovery of deletions and duplications, even on the microscopic scale which may lead to the identification of candidate genes to be further explored by other cytological techniques.Through the use of DNA microarrays in conjunction with CGH techniques, the more specific form of array CGH (aCGH) has been developed, allowing for a locus-by-locus measure of CNV with increased resolution as low as 100 kilobases. This improved technique allows for the aetiology of known and unknown conditions to be discovered.