What`s the Big Deal About DNA?
... What is a double helix? What do the letters A, T, C, and G stand for? ...
... What is a double helix? What do the letters A, T, C, and G stand for? ...
Document
... FISH Probes • Chromosome‐specific centromere probes (CEP®) – Hybridize to centromere region – Detect aneuploidy in interphase and metaphase ...
... FISH Probes • Chromosome‐specific centromere probes (CEP®) – Hybridize to centromere region – Detect aneuploidy in interphase and metaphase ...
24 October - web.biosci.utexas.edu
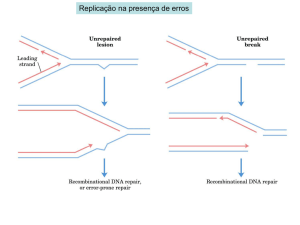
... discussion sections or on next Monday's class no later than 12:00PM. Email attachments and late delivery are not acceptable. 1. What factors ensure the fidelity of replication during DNA synthesis? 2. Define “promoter” and discuss the common features of bacterial promoters. 3. Describe functions of ...
... discussion sections or on next Monday's class no later than 12:00PM. Email attachments and late delivery are not acceptable. 1. What factors ensure the fidelity of replication during DNA synthesis? 2. Define “promoter” and discuss the common features of bacterial promoters. 3. Describe functions of ...
Histones
... complement. The metaphase chromosomes are treated with trypsin (to partially digest the chromosome) and stained with Giemsa. Dark bands that take up the stain are strongly A,T rich (gene poor). The reverse of G-bands is obtained in R-banding. Banding can be used to identify chromosomal abnormalities ...
... complement. The metaphase chromosomes are treated with trypsin (to partially digest the chromosome) and stained with Giemsa. Dark bands that take up the stain are strongly A,T rich (gene poor). The reverse of G-bands is obtained in R-banding. Banding can be used to identify chromosomal abnormalities ...
12711_2011_2534_MOESM1_ESM
... errors. Blank extractions and several negative PCR controls should be performed alongside extractions and amplifications from ancient material. In fact, the quantity of DNA 2 contamination present in laboratory reagents may be so small that it is detected only sporadically in negative controls. Repe ...
... errors. Blank extractions and several negative PCR controls should be performed alongside extractions and amplifications from ancient material. In fact, the quantity of DNA 2 contamination present in laboratory reagents may be so small that it is detected only sporadically in negative controls. Repe ...
Seeking an Increasingly Explicit Definition of Heredity
... Coined the term DNA fingerprinting and was the first to use DNA polymorphisms in paternity, immigration, and murder cases. National Center for Human Genome Research created. $3 billion dollar effort to sequence human genome. 1990 Project launched ...
... Coined the term DNA fingerprinting and was the first to use DNA polymorphisms in paternity, immigration, and murder cases. National Center for Human Genome Research created. $3 billion dollar effort to sequence human genome. 1990 Project launched ...
DNA Technology
... use one of the examples listed above or find your own. Be specific in explaining how the technique was used. Cite your sources – not the textbook. This is the major part of your report. DO NOT USE INSULIN or INDENTIFYING CRIMINALS as examples. Find something less common. 3. If this is a controversia ...
... use one of the examples listed above or find your own. Be specific in explaining how the technique was used. Cite your sources – not the textbook. This is the major part of your report. DO NOT USE INSULIN or INDENTIFYING CRIMINALS as examples. Find something less common. 3. If this is a controversia ...
Microbiology Unit 3 Study Guide
... 5. Which enzyme makes RNA by reading a strand of DNA? 6. Which enzymes cut DNA in specific locations? 7. What occurs during transcription? 8. What are the steps to obtaining DNA fragments for gel electrophoresis? 9. Which enzyme reads DNA to make a new copy of DNA? 10. How has Escherichia coli been ...
... 5. Which enzyme makes RNA by reading a strand of DNA? 6. Which enzymes cut DNA in specific locations? 7. What occurs during transcription? 8. What are the steps to obtaining DNA fragments for gel electrophoresis? 9. Which enzyme reads DNA to make a new copy of DNA? 10. How has Escherichia coli been ...
MolecularBiology1APLab6
... • Contain random DNA fragments that are collected or exchanged w/ other bacteria • Contain nonsense information • Sometimes contain useful information like antibiotic resistance ...
... • Contain random DNA fragments that are collected or exchanged w/ other bacteria • Contain nonsense information • Sometimes contain useful information like antibiotic resistance ...
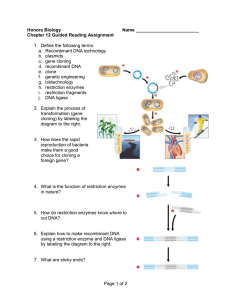
5echap12guidedreading
... 3. How does the rapid reproduction of bacteria make them a good choice for cloning a foreign gene? ...
... 3. How does the rapid reproduction of bacteria make them a good choice for cloning a foreign gene? ...
the element makes na RNA copy of itself which is reversed
... meiosis I, general recombination in bacteria, and viral genetic exchange. • Molecular mechanism proposed by Holliday and Whitehouse (1964). • Depends on complementary base pairing. ...
... meiosis I, general recombination in bacteria, and viral genetic exchange. • Molecular mechanism proposed by Holliday and Whitehouse (1964). • Depends on complementary base pairing. ...
DNA Structure powerpoint
... • Why is DNA wrapped so tightly? • How are DNA, proteins, and traits related? ...
... • Why is DNA wrapped so tightly? • How are DNA, proteins, and traits related? ...
AT CG - Middletown Public Schools
... DNA and Mutations DNA is made up of nucleotides that each contain a sugar, a phosphate, and a base. The four possible bases are adenine, cytosine, thymine, and guanine. Remember that adenine and thymine are complementary and form pairs, and cytosine and guanine are complementary and form pairs. 1. B ...
... DNA and Mutations DNA is made up of nucleotides that each contain a sugar, a phosphate, and a base. The four possible bases are adenine, cytosine, thymine, and guanine. Remember that adenine and thymine are complementary and form pairs, and cytosine and guanine are complementary and form pairs. 1. B ...
What are the three steps in PCR?
... It is often used in DNA fingerprinting It requires gel electrophoresis which separates DNA by size ...
... It is often used in DNA fingerprinting It requires gel electrophoresis which separates DNA by size ...
Some No-Nonsense Facts on
... Teosinte has been selectively bred since Genes undergo mutation when 8000BC. Teosinte has been genetically their DNA sequence changes. modified to produce more kernals of corn on a larger stalk. Teosinte is now called maize . Maize is used as feed corn for livestock. ...
... Teosinte has been selectively bred since Genes undergo mutation when 8000BC. Teosinte has been genetically their DNA sequence changes. modified to produce more kernals of corn on a larger stalk. Teosinte is now called maize . Maize is used as feed corn for livestock. ...
Bio07_TR__U04_CH13.QXD
... Completion On the lines provided, complete the following sentences. 1. In _______________________ , only animals with desired characteristics are allowed to produce the next generation. 2. Crossing dissimilar individuals to bring together the best of both Organisms is called ________________________ ...
... Completion On the lines provided, complete the following sentences. 1. In _______________________ , only animals with desired characteristics are allowed to produce the next generation. 2. Crossing dissimilar individuals to bring together the best of both Organisms is called ________________________ ...
Comparative genomic hybridization

Comparative genomic hybridization is a molecular cytogenetic method for analysing copy number variations (CNVs) relative to ploidy level in the DNA of a test sample compared to a reference sample, without the need for culturing cells. The aim of this technique is to quickly and efficiently compare two genomic DNA samples arising from two sources, which are most often closely related, because it is suspected that they contain differences in terms of either gains or losses of either whole chromosomes or subchromosomal regions (a portion of a whole chromosome). This technique was originally developed for the evaluation of the differences between the chromosomal complements of solid tumor and normal tissue, and has an improved resoIution of 5-10 megabases compared to the more traditional cytogenetic analysis techniques of giemsa banding and fluorescence in situ hybridization (FISH) which are limited by the resolution of the microscope utilized.This is achieved through the use of competitive fluorescence in situ hybridization. In short, this involves the isolation of DNA from the two sources to be compared, most commonly a test and reference source, independent labelling of each DNA sample with a different fluorophores (fluorescent molecules) of different colours (usually red and green), denaturation of the DNA so that it is single stranded, and the hybridization of the two resultant samples in a 1:1 ratio to a normal metaphase spread of chromosomes, to which the labelled DNA samples will bind at their locus of origin. Using a fluorescence microscope and computer software, the differentially coloured fluorescent signals are then compared along the length of each chromosome for identification of chromosomal differences between the two sources. A higher intensity of the test sample colour in a specific region of a chromosome indicates the gain of material of that region in the corresponding source sample, while a higher intensity of the reference sample colour indicates the loss of material in the test sample in that specific region. A neutral colour (yellow when the fluorophore labels are red and green) indicates no difference between the two samples in that location.CGH is only able to detect unbalanced chromosomal abnormalities. This is because balanced chromosomal abnormalities such as reciprocal translocations, inversions or ring chromosomes do not affect copy number, which is what is detected by CGH technologies. CGH does, however, allow for the exploration of all 46 human chromosomes in single test and the discovery of deletions and duplications, even on the microscopic scale which may lead to the identification of candidate genes to be further explored by other cytological techniques.Through the use of DNA microarrays in conjunction with CGH techniques, the more specific form of array CGH (aCGH) has been developed, allowing for a locus-by-locus measure of CNV with increased resolution as low as 100 kilobases. This improved technique allows for the aetiology of known and unknown conditions to be discovered.